Toronto
From 2007.igem.org
(→REVIEW OF PERTINENT LITERATURE) |
|||
| (30 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | [[Image:blue genes logo.jpg|frame|center]] | |
| - | [ | + | |
| - | == Project: | + | == iGEM Project: Bacterial Neural Network == |
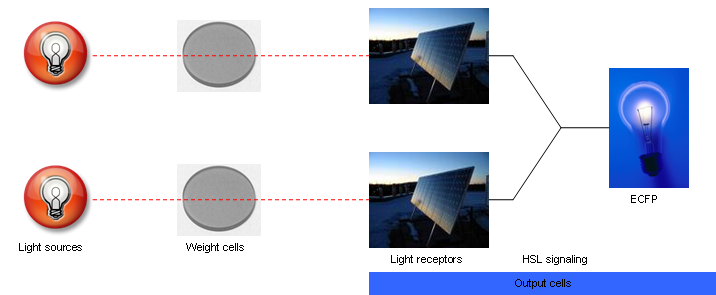
| - | + | [[Image:Fig2.png|thumb|A visual abstraction of the system. Red light inputs pass through filter cells (grey in this image) and hit photoreceptors on the reporter cells (depicted by solar panels). The reporters sum multiple inputs and fluoresce (blue lightbulb).]] | |
| - | + | Our project aims to build a bacterial (E. coli) neural network composed of two cell types, where filter cells (type A) modulate input to reporter cells (type B). The first cell type is stimulated with red light of a specific intensity and duration, and will turn blue in proportion to that "pulse" of light. Populations of type A will be physically mounted above those of type B, acting as a light filter. Type B cells are receptive to the same wavelength of light, and will fluoresce in proportion to the amount of light they receive. | |
| - | + | Neural networks are unique in that their ability to signal both forward (type A influencing type B) and backward (type B influencing type A) results in the ability to learn. Training sessions can be performed with predefined inputs and outputs, and repeated iterations will increase the probability that giving the network an input will produce the desired output. Our neural network training functionality will be implemented through cell-cell signalling from type B cells to type A cells, adjusting the strength of the light filter. | |
| - | + | Depending on the training strategy used, our neural network can learn to function as either an AND or an OR gate. Essentially, our neural network will be able to sum a number of inputs and provide a proportionate output. Once training is completed with a few inputs, we should be able to provide novel inputs to the network and produce appropriate responses. This is a step towards demonstrating fuzzy logic (as opposed to traditional digital logic) in genetic circuits. | |
| - | + | For a more comprehensive description including genetic circuit diagrams, please see [[Toronto/Project Description|E. Coli Neural Network]]. (Updated proposal!) | |
| - | + | '''Background:''' What is a neural network, anyway? See the [http://en.wikipedia.org/wiki/Neural_network Wikipedia definition of a neural network]. | |
| - | + | == Simulated Models == | |
| + | We are currently working on two types of simulations. | ||
| - | + | === Ordinary Differential Equations === | |
| - | + | <center>[[Image:ODEpic.gif]]</center> | |
| - | + | ||
| - | + | An example of the differential equations used to model the neural network. This set defines the luxR-eCFP pathway in the second cell type. | |
| - | + | === Monte Carlo Stochastic Simulation === | |
| - | + | <center>[[Image:output cell layer.gif]]</center> | |
| - | + | This graph depicts the second cell type responding to light. The blue and purple lines indicate two cell populations producing LuxI, while the red line indicates total HSL production. In our neural network, LuxI is produced in response to light input, and expression of LuxI controls the expression of HSL; therefore, this cell type can "add" the light inputs to produce appropriate HSL output. The x -axis depicts iterations of the simulation, while the y-axis is a measure of output (eg. concentration in the system). | |
| - | + | <center>[[Image:one input output simulation.gif]]</center> | |
| + | This figure demonstrates the predicted behaviour of the complete neural network. The blue indicates the size of the light absorbing cell population (ie. the first cell type, which quickly reaches a steady state. The light yellow line represents a steady red input light, while the orange line shows the amount of light being passed to the second cell type (shown in red in the diagram below the graph). Finally, the red line on the graph shows the final output increasing over successive iterations and then reaching a consistent level over time. | ||
| + | == Construction == | ||
| + | '''Construction strategy:''' Using classical DNA transformation and ligation techniques, we plan to build six testing constructs to provide experimental constants for our simulated models of the neural network. The simulations will then assist us in optimizing our experimental conditions for training the final circuits. Building test constructs also results in reliable intermediate parts that can be used to quickly assemble the complete neural network. | ||
| - | + | [[Toronto/Lab Notebook|Lab Notebook]] - watch our daily wetlab progress online | |
| - | + | ||
| - | + | ||
| - | + | [[Toronto/Design Updates|Design Updates]] - the circuit diagrams for the neural network and testing constructs | |
| - | + | ||
| - | + | ||
| + | Lab schedule - the lab schedule is now obsolete. The most recent version is available at [http://igem.skule.ca/lab/schedule.htm BlueGenes lab schedules]. | ||
| - | + | [[Toronto/Lab Protocols|Lab Protocols]] - online versions of our lab protocols. These can also be found on our static site, igem.skule.ca. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | == The Team == | |
| + | [[Toronto/Team Name|What is BlueGenes?]] - a brief overview of who we are, and where the name comes from (not pants!) | ||
| - | + | [[Toronto/Roster|Meet the Team]] - team roster, photos, and profiles | |
| - | + | iGEM 2006 Wiki: [http://parts2.mit.edu/wiki/index.php/University_of_Toronto_2006 Blue Water]. Of particular note is the lab notebook (click on Construction under Committees). | |
| - | + | == Sponsors == | |
| + | [[Toronto/Sponsors|Sponsors]] | ||
| - | + | If you would like to support us, please go to [http://igem.skule.ca/finance/opportunities.htm Sponsorship] for more on becoming a sponsor. | |
| + | == Miscellany == | ||
| + | '''Website:''' [http://www.igem.skule.ca igem.skule.ca] - other information about BlueGenes can be found here. | ||
| - | + | '''Contact Us:''' igem[at]skule[dot]ca | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ''''' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 03:58, 27 October 2007
Contents |
iGEM Project: Bacterial Neural Network
Our project aims to build a bacterial (E. coli) neural network composed of two cell types, where filter cells (type A) modulate input to reporter cells (type B). The first cell type is stimulated with red light of a specific intensity and duration, and will turn blue in proportion to that "pulse" of light. Populations of type A will be physically mounted above those of type B, acting as a light filter. Type B cells are receptive to the same wavelength of light, and will fluoresce in proportion to the amount of light they receive.
Neural networks are unique in that their ability to signal both forward (type A influencing type B) and backward (type B influencing type A) results in the ability to learn. Training sessions can be performed with predefined inputs and outputs, and repeated iterations will increase the probability that giving the network an input will produce the desired output. Our neural network training functionality will be implemented through cell-cell signalling from type B cells to type A cells, adjusting the strength of the light filter.
Depending on the training strategy used, our neural network can learn to function as either an AND or an OR gate. Essentially, our neural network will be able to sum a number of inputs and provide a proportionate output. Once training is completed with a few inputs, we should be able to provide novel inputs to the network and produce appropriate responses. This is a step towards demonstrating fuzzy logic (as opposed to traditional digital logic) in genetic circuits.
For a more comprehensive description including genetic circuit diagrams, please see E. Coli Neural Network. (Updated proposal!)
Background: What is a neural network, anyway? See the [http://en.wikipedia.org/wiki/Neural_network Wikipedia definition of a neural network].
Simulated Models
We are currently working on two types of simulations.
Ordinary Differential Equations
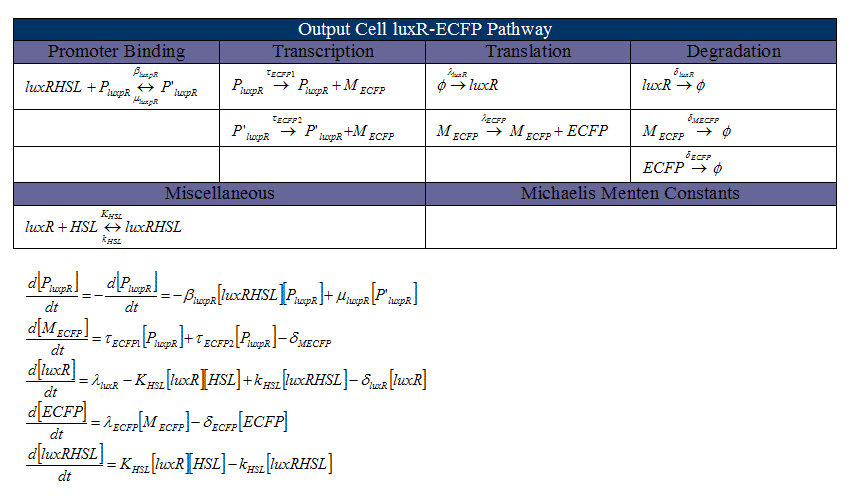
An example of the differential equations used to model the neural network. This set defines the luxR-eCFP pathway in the second cell type.
Monte Carlo Stochastic Simulation
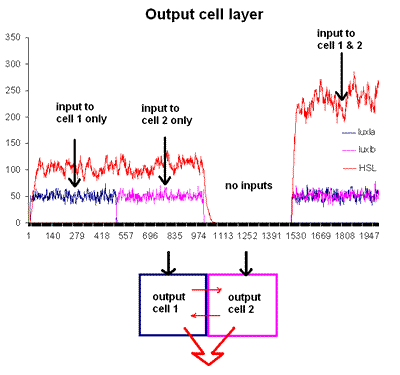
This graph depicts the second cell type responding to light. The blue and purple lines indicate two cell populations producing LuxI, while the red line indicates total HSL production. In our neural network, LuxI is produced in response to light input, and expression of LuxI controls the expression of HSL; therefore, this cell type can "add" the light inputs to produce appropriate HSL output. The x -axis depicts iterations of the simulation, while the y-axis is a measure of output (eg. concentration in the system).
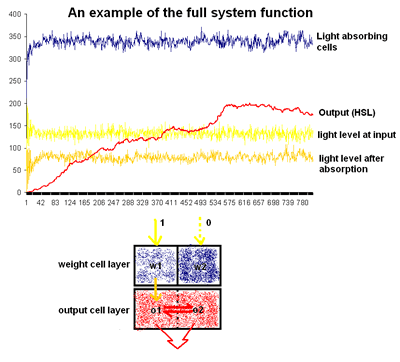
This figure demonstrates the predicted behaviour of the complete neural network. The blue indicates the size of the light absorbing cell population (ie. the first cell type, which quickly reaches a steady state. The light yellow line represents a steady red input light, while the orange line shows the amount of light being passed to the second cell type (shown in red in the diagram below the graph). Finally, the red line on the graph shows the final output increasing over successive iterations and then reaching a consistent level over time.
Construction
Construction strategy: Using classical DNA transformation and ligation techniques, we plan to build six testing constructs to provide experimental constants for our simulated models of the neural network. The simulations will then assist us in optimizing our experimental conditions for training the final circuits. Building test constructs also results in reliable intermediate parts that can be used to quickly assemble the complete neural network.
Lab Notebook - watch our daily wetlab progress online
Design Updates - the circuit diagrams for the neural network and testing constructs
Lab schedule - the lab schedule is now obsolete. The most recent version is available at [http://igem.skule.ca/lab/schedule.htm BlueGenes lab schedules].
Lab Protocols - online versions of our lab protocols. These can also be found on our static site, igem.skule.ca.
The Team
What is BlueGenes? - a brief overview of who we are, and where the name comes from (not pants!)
Meet the Team - team roster, photos, and profiles
iGEM 2006 Wiki: [http://parts2.mit.edu/wiki/index.php/University_of_Toronto_2006 Blue Water]. Of particular note is the lab notebook (click on Construction under Committees).
Sponsors
If you would like to support us, please go to [http://igem.skule.ca/finance/opportunities.htm Sponsorship] for more on becoming a sponsor.
Miscellany
Website: [http://www.igem.skule.ca igem.skule.ca] - other information about BlueGenes can be found here.
Contact Us: igem[at]skule[dot]ca