Chiba/Engeneering Flagella
From 2007.igem.org
(→Our Aim) |
(→Our Aim) |
||
| (22 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
[[Image:chiba_logo.png|center]] | [[Image:chiba_logo.png|center]] | ||
__NOTOC__ | __NOTOC__ | ||
| - | {| style="border:0;width:100%;" cellpadding="20px" cellspacing="0" | + | {| style="border:0;width:100%;font-family:'Trebuchet MS'" cellpadding="20px" cellspacing="0" |
| align="center" | | | align="center" | | ||
| - | [[Chiba|Introduction]] | [[Chiba/Project_Design|Project Design]] ( [[Chiba/Engeneering_Flagella|1.Affinity Tag]] | [[Chiba/Communication|2.Communication Module]] | [[Chiba/Quorum_Sensing|3.Size Control]] ) | [[Chiba/Making Marimo|Making Marimos]] | [[Chiba/Goal|Our Goal]]<br> | + | [[Chiba|Home]]<br> |
| + | <span style="font-size:120%;font-weight:bold;">[[Chiba/Introduction|Introduction]] | [[Chiba/Project_Design|Project Design]] ( [[Chiba/Engeneering_Flagella|1.Affinity Tag]] | [[Chiba/Communication|2.Communication Module]] | [[Chiba/Quorum_Sensing|3.Size Control]] ) | [[Chiba/Making Marimo|Making Marimos]] | [[Chiba/Goal|Our Goal]]</span><br> | ||
[[Chiba/Acknowledgements|Acknowledgements]] | [[Chiba/Team_Members|Team Members]] | [http://chem.tf.chiba-u.jp/igem/ iGEM Chiba Website] | [[Chiba/Members_Only|メンバ連絡簿]] | [[Chiba/Acknowledgements|Acknowledgements]] | [[Chiba/Team_Members|Team Members]] | [http://chem.tf.chiba-u.jp/igem/ iGEM Chiba Website] | [[Chiba/Members_Only|メンバ連絡簿]] | ||
|} | |} | ||
| - | + | ==Affinity Tags== | |
| - | == | + | |
===Our Aim=== | ===Our Aim=== | ||
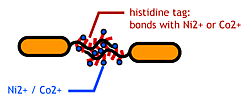
| - | [[Image:Chiba_stickbacteria.png|frame|'''Fig. 9''' | + | [[Image:Chiba_stickbacteria.png|frame|'''Fig. 9''' Affinity tags.]] |
| - | To make | + | To make affinity tags on ''E.coli'', we focused on their flagella that are located outside the cells. We used the following mechanisms: |
*Display sticky peptides in flagellar filament. | *Display sticky peptides in flagellar filament. | ||
*His-tag. The imidazole group in histidines make a complex with metal ions. | *His-tag. The imidazole group in histidines make a complex with metal ions. | ||
| Line 36: | Line 36: | ||
*We inserted the short peptide with six histidine (“His-Tag”) into the fliC D3 domain. | *We inserted the short peptide with six histidine (“His-Tag”) into the fliC D3 domain. | ||
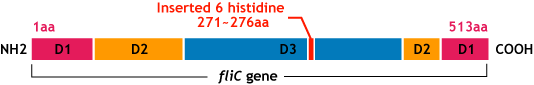
| - | [[Image:Chiba_flichisgene.png]]<br> | + | [[Image:Chiba_flichisgene.png|frame|'''Fig.10''' flic his gene]]<br> |
*[[Image:Chiba_check.png]] Sequence Confirmed<br> | *[[Image:Chiba_check.png]] Sequence Confirmed<br> | ||
| Line 56: | Line 56: | ||
====Testing Procedure==== | ====Testing Procedure==== | ||
| - | #pUC19-FliC-His was transformed to JW1908(''fliC'') and GI826(''fliC'' ''motB''). | + | #pUC19-FliC-His was transformed to JW1908(''fliC''), JW0747(''MotB''), and GI826(''fliC'' ''motB''). |
#Grown to stationary phase | #Grown to stationary phase | ||
#Culture suspended with Dynabeads (Metal-IDA), allowing to the affinity adsorption | #Culture suspended with Dynabeads (Metal-IDA), allowing to the affinity adsorption | ||
| Line 65: | Line 65: | ||
====Results&Discussion==== | ====Results&Discussion==== | ||
'''1.''Stickiness'' check using FliC strain''' | '''1.''Stickiness'' check using FliC strain''' | ||
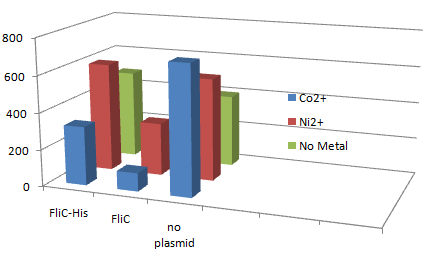
| - | [[Image:Chiba Beads-Adsorption result3.png|frame|left| | + | [[Image:Chiba Beads-Adsorption result3.png|frame|left|'''Fig. 11''' Binding test using Strain JW1908(''⊿fliC'',).]]<br clear="all"> |
*Cell without His-FliC bound better to the Beads? No way! | *Cell without His-FliC bound better to the Beads? No way! | ||
*We thought the problem might be the super-fast revolution of flagella itself. We decided to try MotB strain.<br><br><br> | *We thought the problem might be the super-fast revolution of flagella itself. We decided to try MotB strain.<br><br><br> | ||
| - | [[Image: | + | '''2.''Stickiness'' check using MotB strain''' |
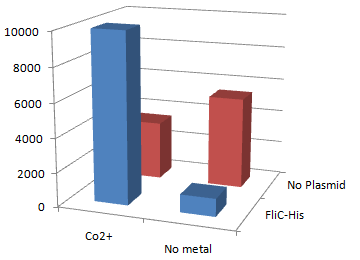
| - | * | + | [[Image:Chiba Beads-Adsorption result2.png|frame|left|'''fig. 12''' Binding test using Strain JW0747(''⊿motB'').]]<br clear="all"> |
| - | Co<sup>2+</sup> | + | *Mot B deletion provides cell with the flagella completely assembled but not rotating. <br> |
| - | *In the presence of FliC-His, cobalt ion adsorb bacteria stronger than nickel ion. | + | *'''This time everything worked out!''' Only in the presence of Co<sup>2+</sup>Bacteria with His-FliC sticked to the Co-IDA beads very well. |
| - | + | *In this strain, FliC-His is assembled with wildtype FliC coded in genomic DNA. Nevertheless, the binding efficiency was at the same level (not shown). it seems that His-Tag displayed on the flagella is enough to do its work. | |
| - | + | *On the other hand, the deletion of MotB turned out to be vital for sticking the tagged flagella together. | |
| - | + | *In the presence of FliC-His, cobalt ion adsorb bacteria stronger than nickel ion, this was more or less the expected result. | |
| + | |||
| + | |||
<!-- | <!-- | ||
| Line 83: | Line 85: | ||
**Beadsとヒスチジンの反応では、Co2+が中心に位置し、ヒスチジンの空間的位置に対する要求性が厳密になっています。連続するヒスチジンや空間的に適切に配置する隣接ヒスチジンのみがこの反応中心でコバルトに結合します。Ni2+ではこのような空間的要求性はあまり厳密ではありません。このため、Ni2+つきのBeadsを用いた場合、His タグ融合タンパク質以外に存在するヒスチジンも結合してしまいます。 | **Beadsとヒスチジンの反応では、Co2+が中心に位置し、ヒスチジンの空間的位置に対する要求性が厳密になっています。連続するヒスチジンや空間的に適切に配置する隣接ヒスチジンのみがこの反応中心でコバルトに結合します。Ni2+ではこのような空間的要求性はあまり厳密ではありません。このため、Ni2+つきのBeadsを用いた場合、His タグ融合タンパク質以外に存在するヒスチジンも結合してしまいます。 | ||
--> | --> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==References== | ==References== | ||
Latest revision as of 05:26, 27 October 2007
|
Home |
Affinity Tags
Our Aim
To make affinity tags on E.coli, we focused on their flagella that are located outside the cells. We used the following mechanisms:
- Display sticky peptides in flagellar filament.
- His-tag. The imidazole group in histidines make a complex with metal ions.
We combined these two and made a His-tagged flagella in the hope to stick them together via metal ions.
[http://www.npn.jst.go.jp/index.html About flagella]
E.Coli have 5-10 flagella. The flagella is used for swimming and for chemotaxis; the bacteria run when they find attractant, tumble when there is a repellent.
E.coli flagella consist of three parts: a basal body, a hook, and a filament. The filament of E.Coli is a rigid, helical, and cylindrical structure which is 10-15μm long and 23nm thick in diameter. It is built from ~20000 subunits of a ~55kDa single protein, FliC. FliC has three domains, D1,D2,D3; although D1 and D2 are needed for the formation of the functional flagellar filament, D3 domain which sticks outside of the fillament are not essential[3].
"Variable" FliC D3 domain
It is reported that the proteins up to 49.4kDa could be displayed on the cell surface of E.Coli using flagellin fusion protein.[4]
About Histidine Tag
See [http://en.wikipedia.org/wiki/His-tag wikipedia article].
Experiments
Making FliC-his gene
- We inserted the short peptide with six histidine (“His-Tag”) into the fliC D3 domain.
Checking the "Stickiness": Beads Adsorption
Purpose
Confirm that the his-tags are displaied on the flagella and are capable of binding to Co2+- or Ni2+- surface.
Samples
- ⊿fliC strain(JW1908 in KEIO collections [5]) transformed with
- pUC19-fliC-his
- no plasmid
- ⊿fliC,⊿motB strain(GI826)transformed with
- pUC19-fliC-his
- no plasmid
Testing Procedure
- pUC19-FliC-His was transformed to JW1908(fliC), JW0747(MotB), and GI826(fliC motB).
- Grown to stationary phase
- Culture suspended with Dynabeads (Metal-IDA), allowing to the affinity adsorption
- Beads washed with a phosphate buffer (x4)
- E" coli" detached from beads by adding imidazole then spreaded on agar plates.
- The number of the colonies on resultant plates.
Results&Discussion
1.Stickiness check using FliC strain
- Cell without His-FliC bound better to the Beads? No way!
- We thought the problem might be the super-fast revolution of flagella itself. We decided to try MotB strain.
2.Stickiness check using MotB strain
- Mot B deletion provides cell with the flagella completely assembled but not rotating.
- This time everything worked out! Only in the presence of Co2+Bacteria with His-FliC sticked to the Co-IDA beads very well.
- In this strain, FliC-His is assembled with wildtype FliC coded in genomic DNA. Nevertheless, the binding efficiency was at the same level (not shown). it seems that His-Tag displayed on the flagella is enough to do its work.
- On the other hand, the deletion of MotB turned out to be vital for sticking the tagged flagella together.
- In the presence of FliC-His, cobalt ion adsorb bacteria stronger than nickel ion, this was more or less the expected result.
References
3. Kuwajima, G. et al.: J. Bacteriol., 170, 3305-3309 (1988)
4. Ezaki, S. et. al.: J. Ferment. Bioeng., 86, 500-503 (1998)
5. Baba, T. et. al.: Mol. Systems. Biol., 21, 1-10 (2006)