Southern Utah
From 2007.igem.org
(→BioBricks) |
|||
| (24 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<br> | <br> | ||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
[[Southern Utah/Project Description]] | [[Southern Utah/Project Description]] | ||
<p style="margin-bottom: 0in;" align="left"><font size="3"> Southern Utah | <p style="margin-bottom: 0in;" align="left"><font size="3"> Southern Utah | ||
| Line 37: | Line 33: | ||
---- | ---- | ||
| - | Last Updated: | + | Last Updated: 10/22/07 |
| - | '''Team Advisors | + | == Goal == |
| + | <br> | ||
| + | [[Image:CyanideBiosensorGoal.jpg]] | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> We want to make bacteria that glow in response to cyanide.</font></p> | ||
| + | |||
| + | |||
| + | == Motivation == | ||
| + | |||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> Cyanide is a toxic compound found in some water. The most common ways for humans to come in contact with cyanide are through industrial waste, or through the root crop cassava found in the tropics and subtropics. Traditional chemical tests designed to test for the presence of cyanide can be very time consuming and labor intensive. One common technique for testing for cyanide involves titrating a cyanide standard to determine its concentration. This is necessary because cyanide is volatile and the concentration of the standard will change over time. Next, the analyst must perform a distillation with acid and a sodium hydroxide gas trap. This step is necessary to separate complexed cyanide from any metals. For the last step, a series of chemicals is generally added to the sample which give it a color. The intensity of the color is then read with a spectrophotometer. This whole process can take a great deal of time. It may be useful to design bacteria that could detect and signal the presence of cyanide to save time and/or labor.</font></p> | ||
| + | |||
| + | |||
| + | == Past Cyanide Biosensors == | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> There have been several research groups that have already characterized some cyanide biosensors (1 Nakamura and Karube 2003). Most of these involve measuring the inhibition of respiration. Cyanide interferes with the proper functioning of cytochrome C in the respiratory pathway. As a result, less oxygen is consumed and less carbon dioxide is produced by bacteria in the presence of cyanide. Although this method has been shown to be able to detect cyanide concentrations in a linear manner, this indirect process is not very selective. Other compounds such as pesticides and herbicides also decrease the amount of bacterial respiration.</font></p> | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> Lee et al. developed a biosensor using the bacterial strain Pseudomonas fluorescens NCIMB 11764. The P. fluorescens strain can use cyanide as its sole nitrogen source. When this strain biodegrades cyanide it produces cyanate, which is then broken down to ammonia and carbon dioxide. The biosensors employed by Lee et al. measured the byproducts of these reactions involved with the degradation of cyanide. While this method is more selective then previous methods, it is still an indirect method of measuring this toxic compound. | ||
| + | </font></p> | ||
| + | |||
| + | == Implementation == | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> We would like to use a strain of bacteria similar to the one used by Lee et al. and modify it to produce a signal in the presence of cyanide. Studies have shown that cyanide metabolism in the Pseudomonas fluorescens strain is an inducible process (2 Kunz, Nagappan, Silva-Avalos, and Delong 1992). One of the key enzymes that interacts with cyanide in the Pseudomonas fluorescens NCIMB 11764 strain is cyanide nitrilase (3 Fernandez, Dolghih, and Kunz 2004). When we did a search for this enzyme on Pubmed, a version of this enzyme in Pseudomonas fluorescens PfO-1 was one of the best search results returned. Upon further investigation, we found that the Pseudomonas fluorescens PfO-1 strain has had its whole genome sequenced whereas the Pseudomonas fluorescens NCIMB 11764 strain had not. For these reasons, we decided to work with the P. fluorescens PfO-1 strain in our project. </font></p> | ||
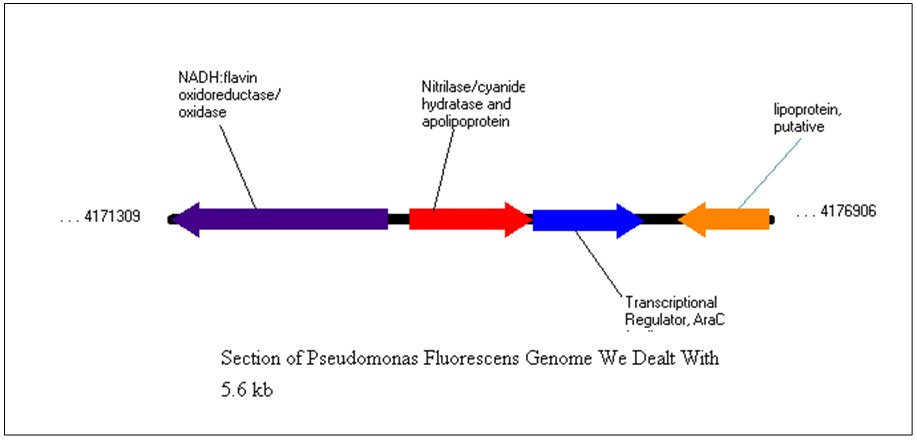
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> The gene that we focused on was a gene that coded for a predicted protein called Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase with the protein ID of YP_349417.1. At first, we didn't realize that there were six other cyanide nitrilase genes in the genome besides this one. Nevertheless, we felt that our first choice was sufficient for this project. Downstream of the cyanide nitrilase gene, there was a gene for a transcriptional regulator of the AraC family. We felt that there was a good possibility that this transcriptional regulator helped control the transcription of the cyanide nitrilase gene. The section of the genome that we focused on is shown below.</font></p> | ||
| + | [[Image:SectionofPseudomonasGenome.jpg]] | ||
| + | |||
| + | |||
| + | == BioBricks == | ||
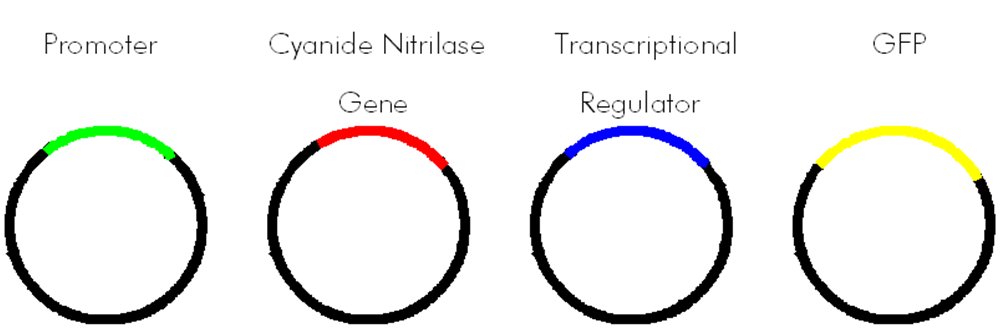
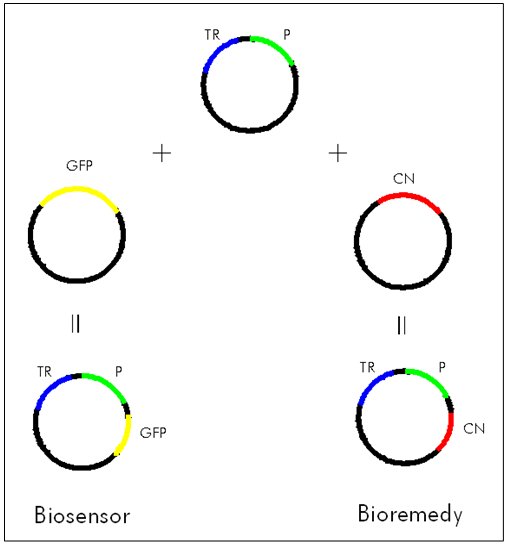
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> Our plan was to isolate genes from the P. fluorescens genome and build BioBricks out of them. We wanted to make a BioBrick that contained the promoter for the cyanide nitrilase gene, and a BioBrick that contained the transcriptional regulator along with its promoter. Note that we did not know exactly where the promoters for these genes were. We chose to treat the region of DNA about 200-300 base pairs upstream of the gene as if it contained the promoter. The transcriptional regulator and the the cyanide nitrilase promoter could then be combined to make a new BioBrick. This new BioBrick could then be attached to GFP (Green Fluorescent Protein). If the cyanide nitrilase gene truly was regulated, and if we chose the correct transcriptional regulator, then this final BioBrick plasmid should cause bacteria to grow in the presence of cyanide. An alternate BioBrick could be made in which some cyanide degrading enzyme such as cyanide nitrilase is used in place of the GFP. This plasmid could be used for bioremediation.</font></p> | ||
| + | [[Image:CyanideBiosensorBioBrick.jpg]]<br> | ||
| + | Useful Parts:<br> | ||
| + | [[Image:CyanideBiosensorUsefulParts.jpg]]<br> | ||
| + | Final Part:<br> | ||
| + | [[Image:CyanideBiosensorFinallPart.jpg]] | ||
| + | |||
| + | == Initial Plans == | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> Our initial plans were to make sure that the cyanide nitrilase gene truly was regulated. In order to do this, we thought about growing two groups of bacteria in different conditions. One group would be exposed to cyanide, and the other would not. Next we would isolate the mRNA from the cell and use a reverse transcriptase to convert this RNA to DNA (essentially creating a cDNA library). If the cyanide nitrilase gene in question truly was regulated, then we should only be able to observe RNA produced from it in the group grown in the presence of cyanide.</font></p> | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> If the cyanide nitrilase gene did turn out to be an inducible gene, then we would need to find out what it's transcriptional regulator was. This could turn out to be a fairly involved process that I won't go into here. Hopefully at the end of the process though, we would find that the AraC transcriptional regulator downstream of the gene would be the true transcriptional regulator just as we thought.</font></p> | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> Unfortunately we did not have enough time to do either of the two checks mentioned previously. Since the deadline was soon approaching, we decided to just go ahead and try to engineer the working plasmid. We would assume that the cyanide nitrilase gene was regulated and that the gene downstream was the correct transcriptional regulator. If everything worked at the end, then we would know that our assumptions were safe. If things did not work at the end, we would have still learned a lot of techniques. At this point we would need to backtrack.</font></p> | ||
| + | |||
| + | == What we had time for == | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> We were quickly running out of time, and we decided to just make a few assumptions to try to build our cyanide biosensor. We started by transforming some E. coli cells with the GFP plasmid. We used the BioBrick part BBa_E0840 because it contained the GFP gene without the promoter; we wanted to use the cyanide nitrilase promoter instead. Below are some pictures of cells transformed with the GFP BioBrick.</font></p> | ||
| + | [[Image:CyanideBiosensorGFPTransformation.jpg]] | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> Next we isolated the genes that we wanted from the Pseudomonas fluorescens strain by designing appropriate primers. A gel run with the DNA from this PCR is shown below.</font></p> | ||
| + | [[Image:CyanideBiosensorPCRGel.jpg]] | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> Next we tried to cut our genes with the appropriate restriction enzymes, mix, ligate, and then transform some cells with the newly formed DNA. As of this writing, we are still having some trouble getting our transformed cells to actually transform with the ligated DNA. Hopefully we can finish before the jamboree.</font></p> | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> Note that this all took much longer to actually do then to write! Rarely did we have something work the very first time. A lot of these procedures were new to us, but we have learned a lot as we have tried to work on this project.</font></p> | ||
| + | |||
| + | == Limitations of Biosensor == | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> If we did successfully create cyanide sensing bacteria, there would still be some limitations. One limitation is that too much cyanide would probably still kill bacteria, even though the P. fluorescens strain can still use it as its sole nitrogen source. Another limitation is that our design would probably only be able to detect free cyanide. Cyanide that is complexed to metals would probably remain undetected. Growth of bacteria on complexed metal cyanides has been reported though. So it is probably possible to extend our intended system with more genes to include the detection of complexed cyanide.</font></p> | ||
| + | |||
| + | == Conclusions == | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> It is much harder to copy and paste things in biology than on a computer! Our project seemed fairly simple at first. We just wanted to copy a few genes from here and past them over there. Simple enough, right? This process turned out to be much more labor intensive and time consuming than many of the team members imagined at first.</font></p> | ||
| + | |||
| + | |||
| + | == Future Work == | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> There is still much work to be done. We still need to successfully complete building our planned plasmid. If this doesn't work as intended, then we need to determine the inducibility of the cyanide nitrilase gene. Once this is determined, we need to identify the correct transcriptional regulator. In addition to this, another team may desire to extend the functionality of a working cyanide biosensor by allowing it to detect metal complexed cyanides as well.</font></p> | ||
| + | |||
| + | |||
| + | == Acknowledgments == | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"></font></p> | ||
| + | == Citations == | ||
| + | <p style="margin-bottom: 0in;" align="left"><font size="3"> | ||
| + | Citations: | ||
| + | 1. Nakamura, H., Karube, I. (2003). Current research activity in biosensors. Anal Bioanal Chem, 377, 446-468.<br> | ||
| + | 2. Kunz, D.A., Nagappan, O., Silva-Avalos, J., Delong, G.T. (1992). Utilization of Cyanide as a Nitrogenous Substrate by Pseudomonas fluorescens NCIMB 11764: Evidence for Multiple Pathways of Metabolic Conversion. Applied and Environmental Microbiology, 58, 2022-2029.<br> | ||
| + | 3. Fernandez, R.F., Dolghih, E., Kunz, D.A. (2004). Enzymatic Assimilation of Cyanide via Pterin-Dependent Oxygenolytic Cleavage to Ammonia and Formate in Pseudomonas fluorescens NCIMB 11764. Applied and Environmental Microbiology, 70, 121-128.</font></p> | ||
| + | |||
| + | |||
| + | |||
| + | == Team Advisors == | ||
| + | <br> | ||
<u>Dr. Colette Calmelet</u><br> | <u>Dr. Colette Calmelet</u><br> | ||
Associate Professor of Mathematics<br> | Associate Professor of Mathematics<br> | ||
| Line 57: | Line 119: | ||
<br> | <br> | ||
'''Team Members'''<br> | '''Team Members'''<br> | ||
| + | Luke Allen<br> | ||
Kelsey Allen<br> | Kelsey Allen<br> | ||
| + | Stacy Fobert <br> | ||
Vishal Lama<br> | Vishal Lama<br> | ||
| + | Greg Thompson<br> | ||
Ray Uchida<br> | Ray Uchida<br> | ||
Autumn Wassmuth<br> | Autumn Wassmuth<br> | ||
Latest revision as of 19:06, 23 November 2007
Visit our main page here: http://igematsuu.googlepages.com/home
Southern Utah/Project Description
Southern Utah University's iGEM team is developing a cyanide biosensor. This project idea was inspired by the 2006 University of Edinburgh iGEM team which developed an arsenic biosensor. Like arsenic, cyanide is a toxic compound that can contaminate water. Currently, the most common ways for cyanide to come into contact with humans are through industrial wastes and through the root crop cassava. Cassava is a major part of the diet for about 300-500 million people living in the tropics and subtropics.
There are currently already several methods for detecting cyanide in water. However, these methods are time consuming and require many steps. We would like to engineer a strain of bacteria that could produce a signal in response to the presence of cyanide. There are already some strains of bacteria such as Pseudomonas fluorescens PfO-1 that produce enzymes such as nitrilase/cyanide hydratase to degrade cyanide. We believe that the transcription of the genes for these enzymes may be dependent on the presence of cyanide. Therefore, we would like to modify the currently existing genes in the Pseudomonas strain so that GFP is produced in response to this toxic compound instead of the usual cyanide degrading enzymes. A bacterial strain with these capabilities may provide quicker detection of cyanide in the future.
Last Updated: 10/22/07
Contents |
Goal
We want to make bacteria that glow in response to cyanide.
Motivation
Cyanide is a toxic compound found in some water. The most common ways for humans to come in contact with cyanide are through industrial waste, or through the root crop cassava found in the tropics and subtropics. Traditional chemical tests designed to test for the presence of cyanide can be very time consuming and labor intensive. One common technique for testing for cyanide involves titrating a cyanide standard to determine its concentration. This is necessary because cyanide is volatile and the concentration of the standard will change over time. Next, the analyst must perform a distillation with acid and a sodium hydroxide gas trap. This step is necessary to separate complexed cyanide from any metals. For the last step, a series of chemicals is generally added to the sample which give it a color. The intensity of the color is then read with a spectrophotometer. This whole process can take a great deal of time. It may be useful to design bacteria that could detect and signal the presence of cyanide to save time and/or labor.
Past Cyanide Biosensors
There have been several research groups that have already characterized some cyanide biosensors (1 Nakamura and Karube 2003). Most of these involve measuring the inhibition of respiration. Cyanide interferes with the proper functioning of cytochrome C in the respiratory pathway. As a result, less oxygen is consumed and less carbon dioxide is produced by bacteria in the presence of cyanide. Although this method has been shown to be able to detect cyanide concentrations in a linear manner, this indirect process is not very selective. Other compounds such as pesticides and herbicides also decrease the amount of bacterial respiration.
Lee et al. developed a biosensor using the bacterial strain Pseudomonas fluorescens NCIMB 11764. The P. fluorescens strain can use cyanide as its sole nitrogen source. When this strain biodegrades cyanide it produces cyanate, which is then broken down to ammonia and carbon dioxide. The biosensors employed by Lee et al. measured the byproducts of these reactions involved with the degradation of cyanide. While this method is more selective then previous methods, it is still an indirect method of measuring this toxic compound.
Implementation
We would like to use a strain of bacteria similar to the one used by Lee et al. and modify it to produce a signal in the presence of cyanide. Studies have shown that cyanide metabolism in the Pseudomonas fluorescens strain is an inducible process (2 Kunz, Nagappan, Silva-Avalos, and Delong 1992). One of the key enzymes that interacts with cyanide in the Pseudomonas fluorescens NCIMB 11764 strain is cyanide nitrilase (3 Fernandez, Dolghih, and Kunz 2004). When we did a search for this enzyme on Pubmed, a version of this enzyme in Pseudomonas fluorescens PfO-1 was one of the best search results returned. Upon further investigation, we found that the Pseudomonas fluorescens PfO-1 strain has had its whole genome sequenced whereas the Pseudomonas fluorescens NCIMB 11764 strain had not. For these reasons, we decided to work with the P. fluorescens PfO-1 strain in our project.
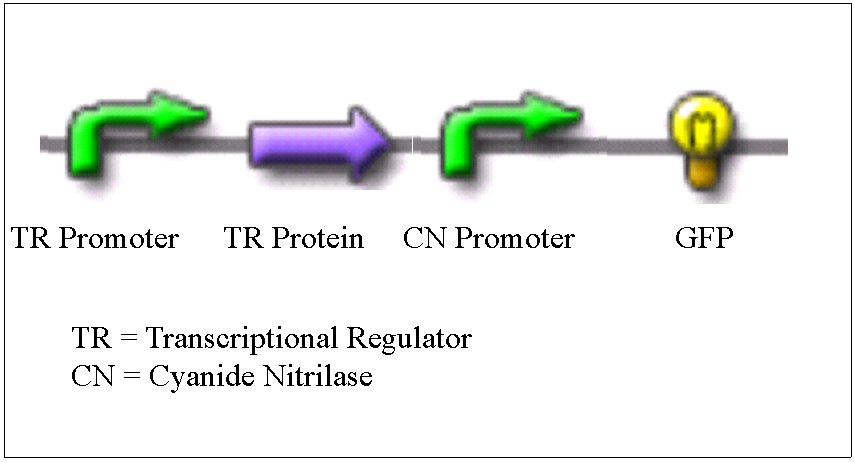
The gene that we focused on was a gene that coded for a predicted protein called Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase with the protein ID of YP_349417.1. At first, we didn't realize that there were six other cyanide nitrilase genes in the genome besides this one. Nevertheless, we felt that our first choice was sufficient for this project. Downstream of the cyanide nitrilase gene, there was a gene for a transcriptional regulator of the AraC family. We felt that there was a good possibility that this transcriptional regulator helped control the transcription of the cyanide nitrilase gene. The section of the genome that we focused on is shown below.
BioBricks
Our plan was to isolate genes from the P. fluorescens genome and build BioBricks out of them. We wanted to make a BioBrick that contained the promoter for the cyanide nitrilase gene, and a BioBrick that contained the transcriptional regulator along with its promoter. Note that we did not know exactly where the promoters for these genes were. We chose to treat the region of DNA about 200-300 base pairs upstream of the gene as if it contained the promoter. The transcriptional regulator and the the cyanide nitrilase promoter could then be combined to make a new BioBrick. This new BioBrick could then be attached to GFP (Green Fluorescent Protein). If the cyanide nitrilase gene truly was regulated, and if we chose the correct transcriptional regulator, then this final BioBrick plasmid should cause bacteria to grow in the presence of cyanide. An alternate BioBrick could be made in which some cyanide degrading enzyme such as cyanide nitrilase is used in place of the GFP. This plasmid could be used for bioremediation.
Initial Plans
Our initial plans were to make sure that the cyanide nitrilase gene truly was regulated. In order to do this, we thought about growing two groups of bacteria in different conditions. One group would be exposed to cyanide, and the other would not. Next we would isolate the mRNA from the cell and use a reverse transcriptase to convert this RNA to DNA (essentially creating a cDNA library). If the cyanide nitrilase gene in question truly was regulated, then we should only be able to observe RNA produced from it in the group grown in the presence of cyanide.
If the cyanide nitrilase gene did turn out to be an inducible gene, then we would need to find out what it's transcriptional regulator was. This could turn out to be a fairly involved process that I won't go into here. Hopefully at the end of the process though, we would find that the AraC transcriptional regulator downstream of the gene would be the true transcriptional regulator just as we thought.
Unfortunately we did not have enough time to do either of the two checks mentioned previously. Since the deadline was soon approaching, we decided to just go ahead and try to engineer the working plasmid. We would assume that the cyanide nitrilase gene was regulated and that the gene downstream was the correct transcriptional regulator. If everything worked at the end, then we would know that our assumptions were safe. If things did not work at the end, we would have still learned a lot of techniques. At this point we would need to backtrack.
What we had time for
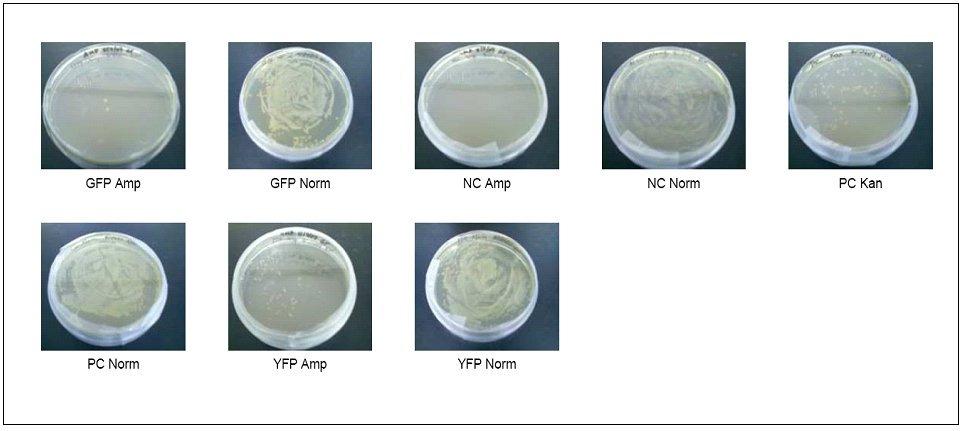
We were quickly running out of time, and we decided to just make a few assumptions to try to build our cyanide biosensor. We started by transforming some E. coli cells with the GFP plasmid. We used the BioBrick part BBa_E0840 because it contained the GFP gene without the promoter; we wanted to use the cyanide nitrilase promoter instead. Below are some pictures of cells transformed with the GFP BioBrick.
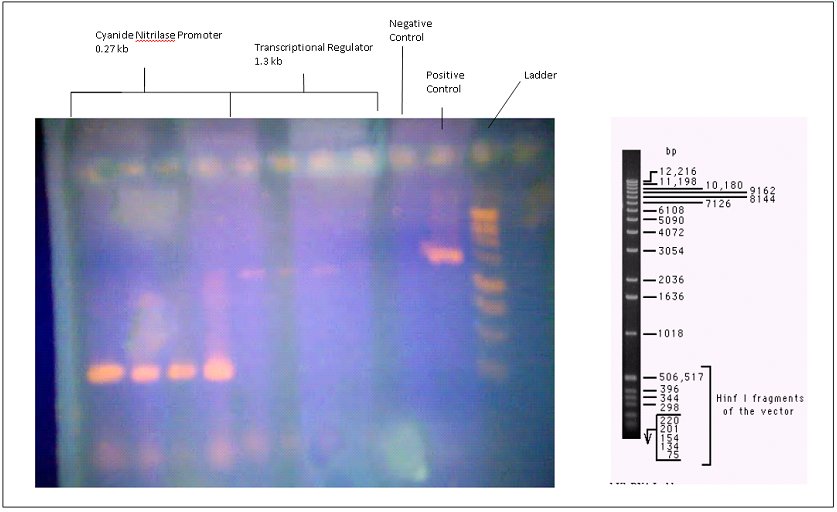
Next we isolated the genes that we wanted from the Pseudomonas fluorescens strain by designing appropriate primers. A gel run with the DNA from this PCR is shown below.
Next we tried to cut our genes with the appropriate restriction enzymes, mix, ligate, and then transform some cells with the newly formed DNA. As of this writing, we are still having some trouble getting our transformed cells to actually transform with the ligated DNA. Hopefully we can finish before the jamboree.
Note that this all took much longer to actually do then to write! Rarely did we have something work the very first time. A lot of these procedures were new to us, but we have learned a lot as we have tried to work on this project.
Limitations of Biosensor
If we did successfully create cyanide sensing bacteria, there would still be some limitations. One limitation is that too much cyanide would probably still kill bacteria, even though the P. fluorescens strain can still use it as its sole nitrogen source. Another limitation is that our design would probably only be able to detect free cyanide. Cyanide that is complexed to metals would probably remain undetected. Growth of bacteria on complexed metal cyanides has been reported though. So it is probably possible to extend our intended system with more genes to include the detection of complexed cyanide.
Conclusions
It is much harder to copy and paste things in biology than on a computer! Our project seemed fairly simple at first. We just wanted to copy a few genes from here and past them over there. Simple enough, right? This process turned out to be much more labor intensive and time consuming than many of the team members imagined at first.
Future Work
There is still much work to be done. We still need to successfully complete building our planned plasmid. If this doesn't work as intended, then we need to determine the inducibility of the cyanide nitrilase gene. Once this is determined, we need to identify the correct transcriptional regulator. In addition to this, another team may desire to extend the functionality of a working cyanide biosensor by allowing it to detect metal complexed cyanides as well.
Acknowledgments
Citations
Citations:
1. Nakamura, H., Karube, I. (2003). Current research activity in biosensors. Anal Bioanal Chem, 377, 446-468.
2. Kunz, D.A., Nagappan, O., Silva-Avalos, J., Delong, G.T. (1992). Utilization of Cyanide as a Nitrogenous Substrate by Pseudomonas fluorescens NCIMB 11764: Evidence for Multiple Pathways of Metabolic Conversion. Applied and Environmental Microbiology, 58, 2022-2029.
3. Fernandez, R.F., Dolghih, E., Kunz, D.A. (2004). Enzymatic Assimilation of Cyanide via Pterin-Dependent Oxygenolytic Cleavage to Ammonia and Formate in Pseudomonas fluorescens NCIMB 11764. Applied and Environmental Microbiology, 70, 121-128.
Team Advisors
Dr. Colette Calmelet
Associate Professor of Mathematics
calmelet@suu.edu
Dr. Bruce Howard
Assistant Professor of Chemistry
howard@suu.edu
435-586-7930
Dr. Charlotte Pedersen
Assistant Professor of Biology
pedersen@suu.edu
435-586-7929
Team Members
Luke Allen
Kelsey Allen
Stacy Fobert
Vishal Lama
Greg Thompson
Ray Uchida
Autumn Wassmuth
Kurt Whittemore
Contact the team at: igematsuu@gmail.com