Paris/Notebook
From 2007.igem.org
(→Friday, July 6) |
|||
| Line 51: | Line 51: | ||
== Friday, July 6 == | == Friday, July 6 == | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | *Making 10 petri dish (LB+ | + | *Preparation of DAP solution from the powder (50mM). See [[Paris/PROTOCOLS | Protocols]] |
| - | + | ||
| - | + | *Making 10 petri dish (LB+tet+citrate+DAP). See [[Paris/PROTOCOLS | Protocols]] | |
| - | * | + | |
| - | + | ||
| - | + | *Making 10 petri dish (LB+erythromycin+citrate+DAP). See [[Paris/PROTOCOLS | Protocols]] | |
| - | + | ||
| + | |||
| + | |||
[[Paris|<<home]] | [[Paris|<<home]] | ||
Revision as of 12:38, 9 July 2007
Contents |
Monday, July 2
First day in the lab !
- Overview of the project
- Planning of the lab work
- Primer design
We got the W121 strain from a lab in Pasteur Institute. This strain is [DapA-; Erythromycin R], but also has a couple of other mutations we are not interested in. Thus we will need to do a transduction of the deletion to the strain we will use: MG1655
We launched ON culture of W121 to prepare a stock of P1 phages.
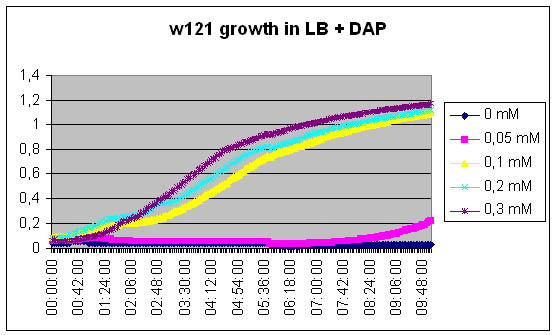
In order to model the dynamics of the synthetic organism, we need to measure several parameters. Two of which are: - The growth of the [DapA-] strain relative to the DAP concentration of the medium; - The excretion of DAP by the [DapA+] strain.
The first one is easy, we just need to measure growth kinetics of our [DapA-] strain contingent on the concentration of the DAP we add in the medium. The second one is more tricky. Direct dosage of DAP is quite complicated and expensive, thus we will try a kind of bio-measurement. We will grow the [DapA+] strain ON, then we will centrifugate to recover the culture medium. We will filter this culture medium to sterilize it and we will grow a [DapA-] strain in it. The growth curve should enable us evaluating the medium concentration in DAP.
Tuesday, July 3
- Preparation of the P1 stock on the w121 strain. See protocols.
- Kinetics measurements:
200ul LB: +0µl DAP, +2µl DAP, +4µl DAP, +8µl DAP, +12µl DAP
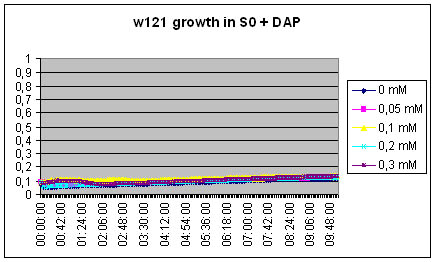
200ul S0: +0µl DAP, +2µl DAP, +4µl DAP, +8µl DAP, +12µl DAP
S0= Centrifuged and filtered medium of MG1655 ON
DAP is 5mM
Wednesday, July 4
- Transduction to MG1655 using the P1 stock made on w121. See protocols.
- Growth kinetics:
The w121 did not grow at all in the S0 medium... Either we did something wrong, or their is a logical explanation ! There might be growth inhibitors in the ON medium, their might be no nutriments left in the medium...
We will redo the experiment but this time, the medium will be recovered from exponential phase culture of MG1655
Thursday, July 5
- Culture of 150 ml of MG1655.
At OD = 0.2 : Centrifugation of 25ml of culture, filtration of the supernatant. ==> S0.2
Same thing at OD= 0.4 , 0.6 and 0.8 , leading to the solutions: S0.2, S0.4, S0.6 and S0.8
- Transduction of the DapA deletion into MG1655 failed... We will redo the stock
Friday, July 6
- Preparation of DAP solution from the powder (50mM). See Protocols
- Making 10 petri dish (LB+tet+citrate+DAP). See Protocols
- Making 10 petri dish (LB+erythromycin+citrate+DAP). See Protocols