GTRs in S oneidensis
From 2007.igem.org
| Line 10: | Line 10: | ||
[[Image:transcriptional_regulator_connectivity.jpg]] | [[Image:transcriptional_regulator_connectivity.jpg]] | ||
| + | |||
| + | [[Boston_University | Back]] | ||
Latest revision as of 20:40, 9 July 2007
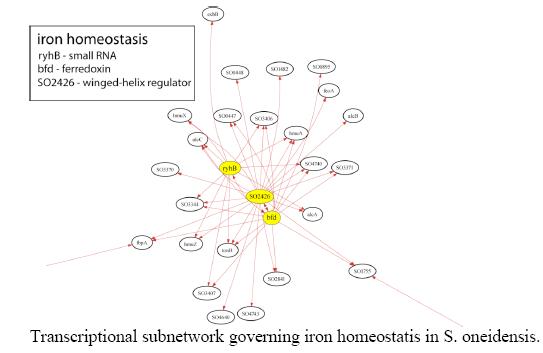
The gram negative bacterial strain Shewanella oneidensis MR-1 was determined to have three global transcriptional regulators; namely hlyU, SO1415 and ttrR. By an algorithm called CLR developed by Dr. Timothy Gardner at Boston University it was determined that the regulation of these transcription factors affected the expression of numerous other genes. Once CLR produces a map, effects of genetic engineering become more predictable, and those modifications leading to the most beneficial result will be easier to identify. The following is an example of a transcriptional network inferred by CLR for just one of S.oneidensis’s metabolic processes:
Transcriptional subnetwork governing iron homeostasis in S. oneidensis.
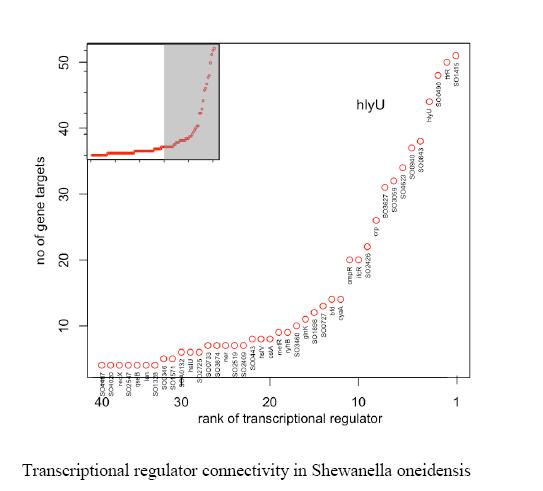
The Gardner Laboratory has used CLR to map 800 regulatory interactions controlling metabolism and electron transport in S. oneidensis. Results from the work have revealed that the transcription regulators SO1415, ttrR, and hlyU affect the largest number of genes involved in these processes [2] as shown in the figure below.
The Gardner Laboratory has used CLR to map 800 regulatory interactions controlling metabolism and electron transport in S. oneidensis. Results from the work have revealed that the transcription regulators SO1415, ttrR, and hlyU affect the largest number of genes involved in these processes [2] as shown in the figure below.