Lethbridge/Notebook
From 2007.igem.org
(→August 14 2007) |
(→October 21 2007) |
||
| (41 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
===July 07 2007=== | ===July 07 2007=== | ||
Attempted Transformation of 4 Biobricks. | Attempted Transformation of 4 Biobricks. | ||
| - | 1. BBa_J5526 - Plate 3 well 6F - RFP | + | 1. BBa_J5526 - Plate 3 well 6F - RFP comp. |
2. BBa_I13522 - Plate 2 well 15H - GFP | 2. BBa_I13522 - Plate 2 well 15H - GFP | ||
3. BBa_R0011 - Plate 1 well 7M - LacI inhibited promoter | 3. BBa_R0011 - Plate 1 well 7M - LacI inhibited promoter | ||
| - | 4. BBa_PO440 - Plate 1 well 21K - | + | 4. BBa_PO440 - Plate 1 well 21K - tet R |
Protocol | Protocol | ||
| - | 1. Add 15uL deionized H2O to well | + | 1. Add 15uL deionized H2O to IGEM plate well |
2. Add 1 uL plasmid to 25uL DH5alpha | 2. Add 1 uL plasmid to 25uL DH5alpha | ||
3. Incubate on ice for 30min | 3. Incubate on ice for 30min | ||
| Line 16: | Line 16: | ||
===July 14 2007=== | ===July 14 2007=== | ||
| - | RFP and | + | RFP and tetR had colonies (RFP comp. had only 1 colony). |
| - | GFP and | + | GFP and Lac pr. did not. |
===July 16 2007=== | ===July 16 2007=== | ||
| - | Repeated transformation of GFP, RFP, and | + | Repeated transformation of GFP, RFP comp., and Lac pr., with addition of a cell concentraion step and more cells/plasmid. (Revisions to protocol in bold.) |
Protocol | Protocol | ||
| Line 32: | Line 32: | ||
8. Plate 100uL on amp+ LB plates, incubate overnight at 37 C | 8. Plate 100uL on amp+ LB plates, incubate overnight at 37 C | ||
| - | Picked colonies from RFP and | + | Picked colonies from RFP comp. and tetR and inoculated amp+ LB broth |
===July 17 2007=== | ===July 17 2007=== | ||
| - | GFP and | + | GFP and Lac pr. had colonies. |
| - | RFP did not. | + | RFP comp. did not. |
| - | Performed plasmid prep on RFP and | + | Performed plasmid prep on RFP and tetR cultures according to QIAprep Miniprep Handbook 2nd Ed. Nov '05 rip out bench protocol for microcentrifuge. |
===July 18 2007=== | ===July 18 2007=== | ||
Ran extracted plasmids on a gel | Ran extracted plasmids on a gel | ||
| - | RFP appears to be too small, and cells do not show red flourescence under UV light. Since part appears not to be working, will build RFP part from 5 subparts. | + | RFP comp. appears to be too small, and cells do not show red flourescence under UV light. Since part appears not to be working, will build RFP part from 5 subparts. |
===July 23 2007=== | ===July 23 2007=== | ||
| Line 49: | Line 49: | ||
-had to redo GFP because accidentally transformed plate 1 well 15H the first time | -had to redo GFP because accidentally transformed plate 1 well 15H the first time | ||
2. BBa_C0012 - plate 1 well 5A - Lac I | 2. BBa_C0012 - plate 1 well 5A - Lac I | ||
| - | 3. BBa_B0034 - plate 1 well 3O - RBS | + | 3. BBa_B0034 - plate 1 well 3O - RBS |
| - | 4. BBa_E1010 - plate 2 well 15M - RFP | + | 4. BBa_E1010 - plate 2 well 15M - RFP basic |
5. BBa_B0010 - plate 2 well 3P - T1 | 5. BBa_B0010 - plate 2 well 3P - T1 | ||
6. BBa_B0012 - plate 1 well 1C - T2 | 6. BBa_B0012 - plate 1 well 1C - T2 | ||
| Line 69: | Line 69: | ||
All transformations worked except LacI. | All transformations worked except LacI. | ||
Picked two colonies from each and cultured in 5mL amp+ LB, incubated overnight at 37 C. | Picked two colonies from each and cultured in 5mL amp+ LB, incubated overnight at 37 C. | ||
| - | |||
| - | |||
===August 14 2007=== | ===August 14 2007=== | ||
| Line 108: | Line 106: | ||
PCR did not work correctly, witnessed only primer dimer and plasmid bands | PCR did not work correctly, witnessed only primer dimer and plasmid bands | ||
| - | Will redo with a much shorter extension time as CheZ is only 640bp | + | Will redo with a much shorter extension time as CheZ is only 640bp |
| + | |||
Will redo with a better size marker and a higher percentage agarose gel | Will redo with a better size marker and a higher percentage agarose gel | ||
| Line 134: | Line 133: | ||
1.5% Agarose gel | 1.5% Agarose gel | ||
| - | noticed contamination in no DNA control, will re-make primers, replace H2O stocks and re-do | + | noticed contamination in no DNA control, will re-make primers, replace H2O stocks and re-do. |
| - | + | band appeared in the correct spot | |
===August 22 2007=== | ===August 22 2007=== | ||
Re-made primers from stock solution | Re-made primers from stock solution | ||
| + | |||
Will modify PCR reaction conditions to take into account the low Tm of the regions that actually bind, and then raise annealing temperature after a few cycles when the primer binding regions has been amplified | Will modify PCR reaction conditions to take into account the low Tm of the regions that actually bind, and then raise annealing temperature after a few cycles when the primer binding regions has been amplified | ||
| Line 159: | Line 159: | ||
10.75uL H2O (optima) | 10.75uL H2O (optima) | ||
1uL template (1/10, 1/100, no DNA control) | 1uL template (1/10, 1/100, no DNA control) | ||
| + | |||
| + | ===August 23 2007=== | ||
| + | 2% Agarose gel of PCR | ||
| + | 1- 50bp ladder (didn't fire?) | ||
| + | 2- Riboswitch - No DNA | ||
| + | 3- Riboswitch | ||
| + | 4- Riboswitch + CheZ - No DNA | ||
| + | 5- Riboswitch + CheZ | ||
| + | 6- CheZ - No DNA | ||
| + | 7- CheZ | ||
| + | |||
| + | [[Image:August23.jpg|150px]] | ||
| + | |||
| + | ===August 27 2007=== | ||
| + | |||
| + | Attempted Transformation of 2 Biobricks using XL1-blue supercompotent cells. | ||
| + | 1. BBa_I13507 - Plate 1 well 16N - RFP sub. | ||
| + | 2. BBa_C0012 - Plate 1 well 5A - LacI (repeat) | ||
| + | |||
| + | Protocol | ||
| + | 1. Add 50uL XL1-blue cells with 0.85uL 2-mercaptoethanol to 15mL falcon tube (prechilled) | ||
| + | 2. Leave on ice for 10 min. | ||
| + | 3. Add 15uL deionized H2O to IGEM plate well | ||
| + | 4. Add 1 uL plasmid to cells | ||
| + | 5. Incubate on ice for 30min | ||
| + | 6. Heat shock without shaking for 45sec at 42 C. | ||
| + | 7. Place on ice for 2min | ||
| + | 8. Add 0.9mL SOC media, preheated to 42 C, incubate for 1hr | ||
| + | 9. Plate 200uL on amp+ LB plates, incubate overnight at 37 C | ||
| + | |||
| + | Ran a digest of Lac pr. and tetR plasmids with the intention of ligating tetR behind Lac pr. | ||
| + | reaction conditions | ||
| + | 1. 2uL NEB 2 stock buffer | ||
| + | 2. 0.2uL stock BSA | ||
| + | 3. 5uL prepped plasmid | ||
| + | 4. 0.5uL of each restriction enzyme (two) | ||
| + | 5. 11.8uL dH2O | ||
| + | Used Pst1 + Spe1 for Lac. pr. and used Pst1 + Xba1 for tetR. | ||
| + | |||
| + | ===August 28 2007=== | ||
| + | |||
| + | RFP sub. transformation worked, but LacI did not. Picked colonies from RFP sub. and incubated in amp/LB overnight at 37 C. | ||
| + | Supervisor informed that supercompotent cells used had been in a thaw accident, repeated LacI and RFP comp. transformations with new supercompotent cells using the same protocol | ||
| + | |||
| + | ===August 29 2007=== | ||
| + | |||
| + | RFP comp. worked (1 colony) but LacI once again did not. picked RFP comp. incubated in amp/LB overnight at 37 C. | ||
| + | |||
| + | Since LacI is not working, we are switching the repressor component of the reporter system. Lambda cI and a lambda cI inhibited promoter will be used instead of the LacI components. | ||
| + | |||
| + | Attempted Transformation of 3 Biobricks using XL1-blue supercompotent cells with same protocol as 27/08/07. | ||
| + | 1. BBa_R0051 - Plate 1 well 9C - lambda cI inhibited promoter | ||
| + | 2. BBa_C0051 - Plate 1 well 5G - lambda cI | ||
| + | 3. BBa_J23119 - PLate 3 well 19A - Strong promoter for riboswitch | ||
| + | |||
| + | ===August 30 2007=== | ||
| + | |||
| + | All three transformations worked, picked two colonies from each and incubated overnight at 37 C in LB/amp | ||
| + | |||
| + | The RFP comp. colony picked on the 29th did not grow, but the plates had been left in the incubator for another night and colonies grew on both the LacI and RFP comp. plate. Probably false positives, but picked two colonies from each and inoculated LB/amp | ||
| + | |||
| + | |||
| + | ===September 3 2007=== | ||
| + | |||
| + | Created a new streak plate of RP1616 (cheZ mutant) to use with Fermentas TransformAid Kit to make cells competant for transformation. | ||
| + | |||
| + | Riboswitch PCR | ||
| + | 37.5uL HF Buffer | ||
| + | 7.5uL dNTP | ||
| + | 80.7uL H2O | ||
| + | 1.89uL Phusion | ||
| + | 7.5uL Forward Primer | ||
| + | 7.5uL Reverse Primer | ||
| + | 2.5uL Template/Reaction --> 3 Reactions | ||
| + | |||
| + | ===September 5 2007=== | ||
| + | |||
| + | Performed pruification of the PCR product of CheZ using Qiagen MinElute Kit for PCR purification. | ||
| + | |||
| + | Started overnight culture of RP1616 to use for transformation of Promoter+Riboswitch+CheZ plasmid as well as a puc19 control. | ||
| + | |||
| + | 10% DNA PAGE Gel of Riboswitch PCR | ||
| + | All three are positive! | ||
| + | |||
| + | [[Image:September5.jpg|150px]] | ||
| + | |||
| + | ===September 5 2007=== | ||
| + | PCR Purification of Riboswitch using Qiagen MinElute Kit for PCR Purification | ||
| + | |||
| + | ===September 12 2007=== | ||
| + | |||
| + | Ran a digest of C0051, B0010, B0012, I13522, R0051, and I13507. | ||
| + | Reaction conditions | ||
| + | 1. 5uL NEB 2 4. 1uL RE #1 | ||
| + | 2. 0.5uL BSA 5. 1uL RE #2 | ||
| + | 3. 5uL template 6. 37.5uL ddH2O | ||
| + | Temp. 37C Overnight | ||
| + | Enzymes used | ||
| + | C0051 - EcoR1 + Spe1 I13522 - Xba1 + Pst1 | ||
| + | B0010 - EcoR1 + Xba1 R0051 - Spe1 + Pst1 | ||
| + | B0012 - Spe1 + Pst1 I13507 - Xba1 + Pst1 | ||
| + | |||
| + | ===September 17 2007=== | ||
| + | [Riboswitch]=200ng/uL after purification | ||
| + | |||
| + | Double digestion of Riboswitch | ||
| + | 5uL DNA | ||
| + | 0.3uL EcoRI | ||
| + | 0.3uL PstI | ||
| + | 1uL NEBuffer 2 | ||
| + | 2.4 uL H2o | ||
| + | 10uL Reaction - should be ~100ng/uL. Will try a sticky end and a blunt end ligation in parallel.' | ||
| + | |||
| + | ===September 18 2007=== | ||
| + | [pUC19] cut with EcorI and PstI is about 40ng/uL. Will use 10uL (400ng) for ligation. | ||
| + | [pUC19] cut with SmaI is about 10ng/uL. Will use 10uL (100ng) for ligation. | ||
| + | |||
| + | Riboswitch ligations into pUC19 | ||
| + | |||
| + | Sticky end | ||
| + | 10uL pUC19 (400ng) | ||
| + | 0.9uL riboswitch | ||
| + | 2uL ligation buffer | ||
| + | 2uL ligase (1U/uL) | ||
| + | 5.1uL H2O | ||
| + | |||
| + | Blunt End | ||
| + | 10uL pUC19 (100ng) | ||
| + | 0.3uL riboswitch | ||
| + | 2uL ligation buffer | ||
| + | 1uL ligase | ||
| + | 0.3uL SmaI | ||
| + | 2uL PEG | ||
| + | 4.4uL H2O | ||
| + | |||
| + | ===September 19 2007=== | ||
| + | PCR purification of newly made riboswitch | ||
| + | |||
| + | Transformation of Riboswitch in pUC19 into Ecoli DH5alpha. Plates contained XGal | ||
| + | |||
| + | Quantification gel of Sept 12 digests. | ||
| + | |||
| + | ===September 21 2007=== | ||
| + | Colony screening PCR of Riboswitch/pUC19 | ||
| + | |||
| + | No. 1-8 - blunt end ligations | ||
| + | No. 9-15 - sticky end ligations | ||
| + | No. 16 and 17 - blue colonies | ||
| + | No. 18 - PCR control (positive) | ||
| + | |||
| + | 36uL 10x buffer | ||
| + | 7.2uL dNTP | ||
| + | 152.64uL H2O | ||
| + | 2.16 uL Taq | ||
| + | 18uL Forward primer (riboswitch primer) | ||
| + | 18uL Reverse primer | ||
| + | 36uL MgSO4 | ||
| + | |||
| + | ===September 23 2007=== | ||
| + | 10% DNA PAGE Gels of Colony screening. | ||
| + | |||
| + | All seem positive - will do a restriction with an enzyme that only cuts in the riboswitch | ||
| + | |||
| + | [[Image:September23a.jpg|150px]] | ||
| + | |||
| + | [[Image:September23b.jpg|150px]] | ||
| + | |||
| + | ===September 30 2007=== | ||
| + | Miniprepped Blunt No's 1,4 and 5, and Stick No's 9, 12, 15 using the QiaQuick spin Kit | ||
| + | |||
| + | Single restriction with SpeI | ||
| + | |||
| + | Mix | ||
| + | 12uL NEBuffer 2 | ||
| + | 1.2 uL SpeI (2U) | ||
| + | 1.2uL BSA | ||
| + | 75.6uL H2O | ||
| + | |||
| + | 5uL DNA per reaction, 6 reactions. | ||
| + | |||
| + | 1% Agarose Gels - loaded 5uL of each sample | ||
| + | |||
| + | Number 1 | ||
| + | 1- 1Kb LAdder (4uL) | ||
| + | 2- riboswitch 1 Unrestricted | ||
| + | 3- " Restricted | ||
| + | 4- riboswitch 4 Unrestricted | ||
| + | 5- " Restricted | ||
| + | 6- riboswitch 5 Unrestricted | ||
| + | 7- " REstricted | ||
| + | 8- riboswitch 9 Unrestricted | ||
| + | 9- " Restricted | ||
| + | 10- riboswitch 12 - Unrestricted | ||
| + | 11 " Restricted | ||
| + | 12 - empty | ||
| + | |||
| + | Number 2 | ||
| + | 1- riboswitch 15- unrestricted | ||
| + | 2- " restricted | ||
| + | 3-6 are other things | ||
| + | 7- 1Kb ladder | ||
| + | |||
| + | |||
| + | [[Image:October1a.jpg|150px]] | ||
| + | |||
| + | [[Image:October1b.jpg|150px]] | ||
| + | |||
| + | ===October 20 2007=== | ||
| + | Double digestion of riboswitch in pUC19 with XbaI and PstI | ||
| + | |||
| + | Master Mix | ||
| + | 8uL PstI | ||
| + | 8uL XbaI | ||
| + | 8uL NEBuffer II | ||
| + | 4uL BSA | ||
| + | 12uL H2O | ||
| + | 5uL DNA in each | ||
| + | |||
| + | [[Image:October20.jpg|150px]] | ||
| + | |||
| + | |||
| + | ===October 21 2007=== | ||
| + | Prepared semisolid plates containing theophylline or caffeine. | ||
| + | Plates were made with LB and 0.25% agar and ampicillin. | ||
| + | 0mM, 0.1mM, 1mM, and 5mM Theophylline and Caffeine plates were made by adding the appropriate | ||
| + | amount of 100mM stock solutions to 25mL of warm semisolid media before pouring plate. | ||
| + | |||
| + | A tube of DH5a cells containing the plasmid with CheZ and the riboswitch was grown overnight in LB with ampicillin. | ||
| + | |||
| + | |||
| + | |||
| + | Samples 1-6 seem to work. Pooled samples 5 and 6 for gel extration. | ||
| + | |||
| + | Preperative restriction | ||
| + | 40uL DNA | ||
| + | 2uL XbaI | ||
| + | 2uL PstI | ||
| + | 8uL NEBufferII | ||
| + | 27uL H2O | ||
| + | 1uL BSA | ||
| + | |||
| + | 80uL total, incubate at 37C for two hours. | ||
| + | |||
| + | Gel extraction using Quaigen kit | ||
| + | |||
| + | 2% Agarose gel to quantify: | ||
| + | |||
| + | Lane 1- 100bp ladder (4uL) | ||
| + | Lane 2 - gel extracted riboswitch (4uL) | ||
| + | |||
| + | [[Image:October22.jpg|150px]] | ||
| + | |||
| + | Concentration appears to be approximately 3.3 ng/uL | ||
| + | |||
| + | Ligation of Riboswitch into strong promoter plasmid. | ||
| + | 3uL vector | ||
| + | 4uL riboswitch insert | ||
| + | 1uL ligase | ||
| + | 1uL T4 DNA ligase buffer | ||
| + | 1uL H2O | ||
| + | Total of 10 uL | ||
| + | |||
| + | ===October 22 2007=== | ||
| + | Transformation of riboswitch/strong promotor plasmid | ||
| + | |||
| + | The plates from October 21 2007 were streaked with an inoculating loop to asses motility when cells were exposed various levels of theophylline and caffeine. | ||
| + | |||
| + | ===October 23 2007=== | ||
| + | Streaks made on October 22 2007 were assessed and the results were not clear. The variability present in initial streak size was larger than any difference in cell migration rate due to motility. On the highest concentration of Theophylline plated, there were a much larger number of satellite colonies formed than any other plates. This may be an indication of motility, but is far from conclusive. | ||
| + | |||
| + | Using the same plates, two drops (1uL and 10uL) of liquid culture were placed, with care to reduce variation in size. Plates were incubated overnight at 37 C. | ||
| + | |||
| + | Picked 20 colonies for PCR colony screening | ||
| + | |||
| + | Master mix | ||
| + | 25uL Forward primer | ||
| + | 25uL Reverse primer | ||
| + | 10uL dNTP | ||
| + | 10X buffer 50uL | ||
| + | 15.1uL H2O in each | ||
| + | Swirl colony in each | ||
| + | |||
| + | [[Image:October25.jpg|150px]] | ||
| + | |||
| + | Miniprepped colonies 5-10. | ||
| + | |||
| + | ===October 24 2007=== | ||
| + | The drops from October 23 2007 were assessed and no variation in size (presumably due to motility) was noticed above the variation present in drop size. This suggests that motility assays will need to be carried out at the microscopic level. | ||
| + | |||
| + | ===October 26 2007=== | ||
| + | Sent 2 pUC19 plasmids containing the riboswitch and CheZ as well as a pUC19 derivative plasmid containing a strong promoter with the riboswitch. | ||
<div style="display:block; text-align:center; border:2px solid #6C5; width:500px; margin:auto; margin-top:3em; padding:5px;"> | <div style="display:block; text-align:center; border:2px solid #6C5; width:500px; margin:auto; margin-top:3em; padding:5px;"> | ||
<p> [[Lethbridge | Main Page]] | [[Lethbridge/Summary | Project Overview]] | [[Lethbridge/Timeline | Timeline]] | [[Lethbridge/Notebook | Lab Notebook]] | [[Lethbridge/Team | Team Members]]</p> | <p> [[Lethbridge | Main Page]] | [[Lethbridge/Summary | Project Overview]] | [[Lethbridge/Timeline | Timeline]] | [[Lethbridge/Notebook | Lab Notebook]] | [[Lethbridge/Team | Team Members]]</p> | ||
</div> | </div> | ||
Latest revision as of 23:15, 26 October 2007
July 07 2007
Attempted Transformation of 4 Biobricks.
1. BBa_J5526 - Plate 3 well 6F - RFP comp. 2. BBa_I13522 - Plate 2 well 15H - GFP 3. BBa_R0011 - Plate 1 well 7M - LacI inhibited promoter 4. BBa_PO440 - Plate 1 well 21K - tet R
Protocol
1. Add 15uL deionized H2O to IGEM plate well 2. Add 1 uL plasmid to 25uL DH5alpha 3. Incubate on ice for 30min 4. Heat shock without shaking for 20sec 5. Place on ice for 2min 6. Add 0.5mL LB, incubate for 1hr 7. Plate 100uL on amp+ LB plates, incubate overnight at 37 C
July 14 2007
RFP and tetR had colonies (RFP comp. had only 1 colony). GFP and Lac pr. did not.
July 16 2007
Repeated transformation of GFP, RFP comp., and Lac pr., with addition of a cell concentraion step and more cells/plasmid. (Revisions to protocol in bold.)
Protocol
1. Add 35uL deionized H2O to well 2. Add 5uL plasmid to 50uL DH5alpha 3. Incubate on ice for 30min 4. Heat shock without shaking for 20sec 5. Place on ice for 2min 6. Add 0.5mL LB, incubate for 1hr 7. Spin down cells gently, remove 350uL supernatant and resuspend (gently!) pellet 8. Plate 100uL on amp+ LB plates, incubate overnight at 37 C
Picked colonies from RFP comp. and tetR and inoculated amp+ LB broth
July 17 2007
GFP and Lac pr. had colonies. RFP comp. did not. Performed plasmid prep on RFP and tetR cultures according to QIAprep Miniprep Handbook 2nd Ed. Nov '05 rip out bench protocol for microcentrifuge.
July 18 2007
Ran extracted plasmids on a gel
RFP comp. appears to be too small, and cells do not show red flourescence under UV light. Since part appears not to be working, will build RFP part from 5 subparts.
July 23 2007
Attempted transformation of 6 biobricks
1. BBa_I13522 - plate 2 well 15H - GFP
-had to redo GFP because accidentally transformed plate 1 well 15H the first time
2. BBa_C0012 - plate 1 well 5A - Lac I
3. BBa_B0034 - plate 1 well 3O - RBS
4. BBa_E1010 - plate 2 well 15M - RFP basic
5. BBa_B0010 - plate 2 well 3P - T1
6. BBa_B0012 - plate 1 well 1C - T2
Protocol
1. Add 15uL deionized H2O to well 2. Add 1 uL plasmid to 25uL DH5alpha 3. Incubate on ice for 30min 4. Heat shock without shaking for 20sec 5. Place on ice for 2min 6. Add 0.5mL LB, incubate for 1hr 7. Spin 13000 rpm for 1min, remove 400uL supernatant and resuspend 7. Plate 100uL on amp+ LB plates, incubate overnight at 37 C
July 24 2007
Note book created (All previous entries are transcribed from paper lab notebook)
All transformations worked except LacI. Picked two colonies from each and cultured in 5mL amp+ LB, incubated overnight at 37 C.
August 14 2007
PCR Amplification of Riboswitch and CheZ
-made a 1/200 dilution of maxiprep of pCheZ template -used spec to find concentration of 163ng/uL -made 1/10 and 1/100 dilutions of this for use as PCR template
Riboswitch PCR (recipe/reaction)
5uL 5x phusion polymerase buffer 1uL dNTP mix 1.25uL 20uM Forward Primer 1.25uL 20uM Reverse Primer 0.5uL Phusion polymerase 11.5uL H2O (optima) 5uL template (1/10, 1/100, no DNA control)
CheZ PCR (recipe/reaction)
5uL 5x phusion polymerase buffer 1uL dNTP mix 1.25uL 20uM Forward Primer 1.25uL 20uM Reverse Primer 0.5uL Phusion polymerase 11.5uL H2O (optima) 5uL template (1/10, 1/100, no DNA control)
Reaction Conditions
1X 98 C 1.5min
35X 98 C 30sec
58 C 45sec
72 C 2.5min
1X 72 C 4min
Ran on 0.8% agarose gel with HindIII marker
PCR did not work correctly, witnessed only primer dimer and plasmid bands
Will redo with a much shorter extension time as CheZ is only 640bp
Will redo with a better size marker and a higher percentage agarose gel
August 20 2007
PCR Amplification of Riboswitch and CheZ
Used same template as August 14
CheZ PCR (recipe/reaction)
5uL 5x phusion polymerase buffer 1uL dNTP mix 1.25uL 20uM Forward Primer 1.25uL 20uM Reverse Primer 0.5uL Phusion polymerase 11.5uL H2O (optima) 5uL template (1/10, 1/100, no DNA control)
New reaction conditions
1X 98 C 1min
35X 98 C 15sec
60 C 20sec
72 C 30min
1X 72 C 4min
1.5% Agarose gel
noticed contamination in no DNA control, will re-make primers, replace H2O stocks and re-do. band appeared in the correct spot
August 22 2007
Re-made primers from stock solution
Will modify PCR reaction conditions to take into account the low Tm of the regions that actually bind, and then raise annealing temperature after a few cycles when the primer binding regions has been amplified
Modified Reaction Conditions
1X 98 C 1min
5X 98 C 20sec
41 C 30sec
72 C 1min
30X 98 C 20sec
60 C 30sec
72 C 1min
1X 72 C 4min
New PCR recipie (only minor changes)
5uL 5x phusion polymerase buffer 1uL dNTP mix 1uL 20uM Forward Primer 1uL 20uM Reverse Primer 0.25uL Phusion polymerase 10.75uL H2O (optima) 1uL template (1/10, 1/100, no DNA control)
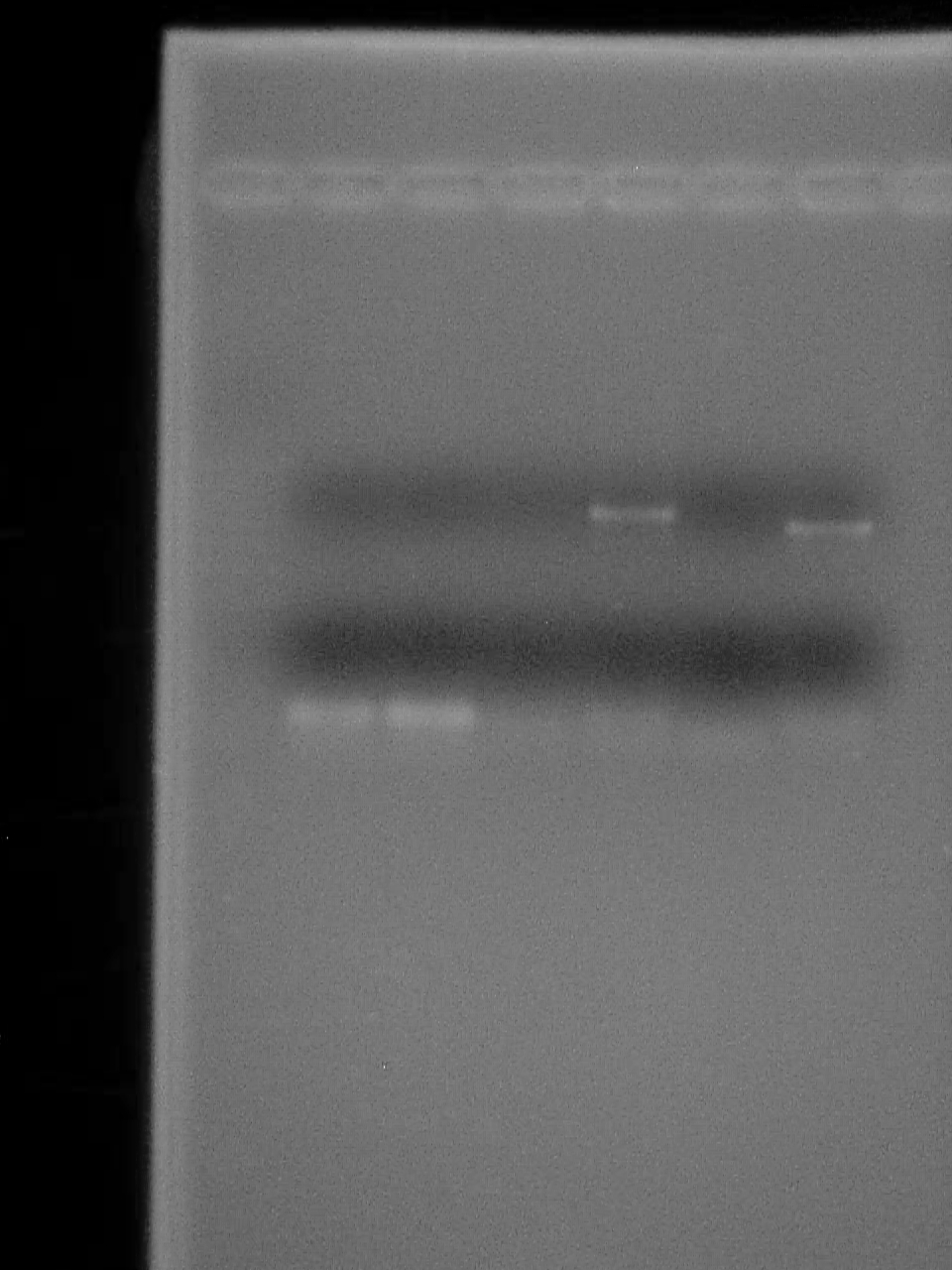
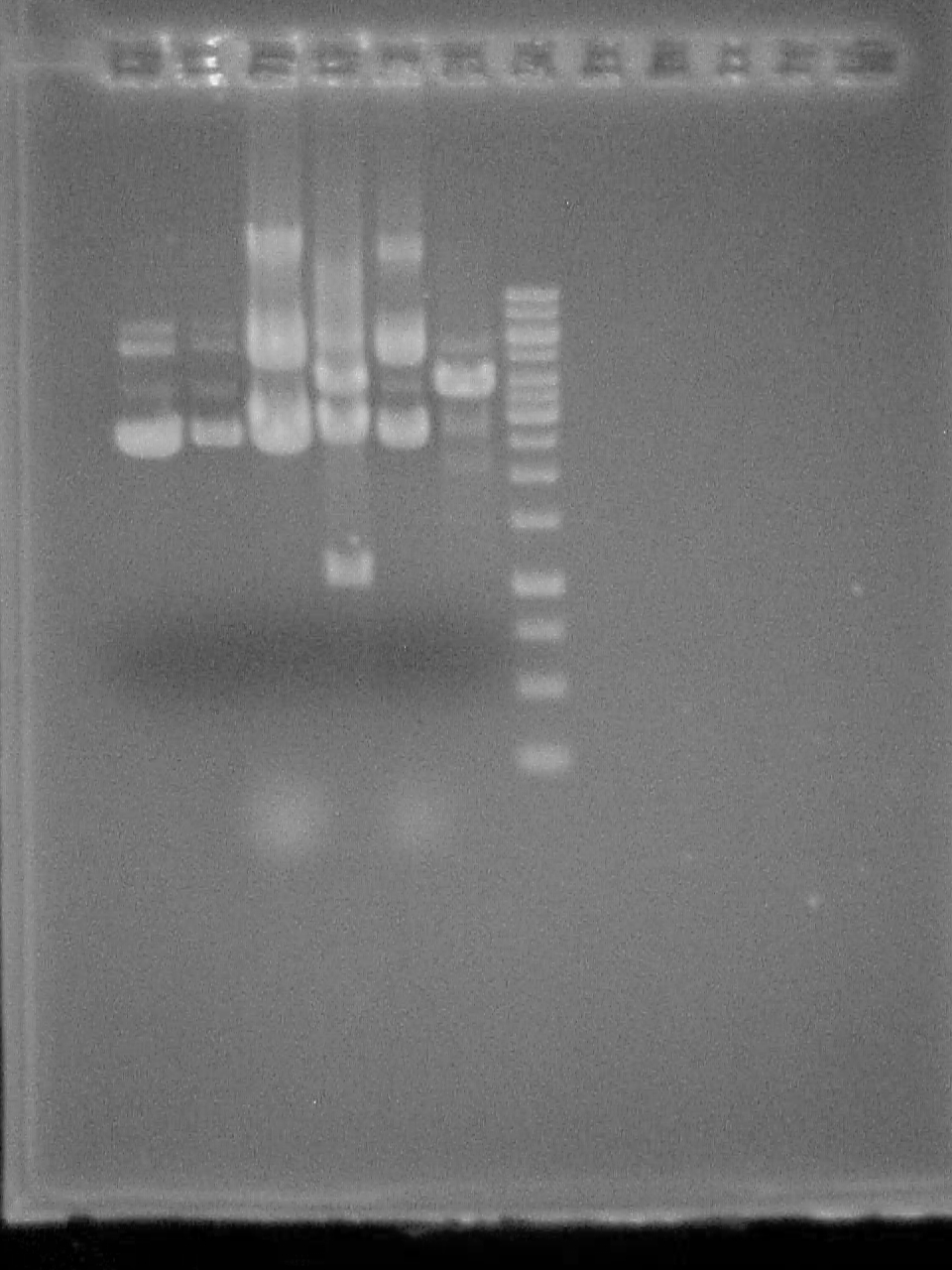
August 23 2007
2% Agarose gel of PCR 1- 50bp ladder (didn't fire?) 2- Riboswitch - No DNA 3- Riboswitch 4- Riboswitch + CheZ - No DNA 5- Riboswitch + CheZ 6- CheZ - No DNA 7- CheZ
August 27 2007
Attempted Transformation of 2 Biobricks using XL1-blue supercompotent cells.
1. BBa_I13507 - Plate 1 well 16N - RFP sub. 2. BBa_C0012 - Plate 1 well 5A - LacI (repeat)
Protocol
1. Add 50uL XL1-blue cells with 0.85uL 2-mercaptoethanol to 15mL falcon tube (prechilled) 2. Leave on ice for 10 min. 3. Add 15uL deionized H2O to IGEM plate well 4. Add 1 uL plasmid to cells 5. Incubate on ice for 30min 6. Heat shock without shaking for 45sec at 42 C. 7. Place on ice for 2min 8. Add 0.9mL SOC media, preheated to 42 C, incubate for 1hr 9. Plate 200uL on amp+ LB plates, incubate overnight at 37 C
Ran a digest of Lac pr. and tetR plasmids with the intention of ligating tetR behind Lac pr.
reaction conditions 1. 2uL NEB 2 stock buffer 2. 0.2uL stock BSA 3. 5uL prepped plasmid 4. 0.5uL of each restriction enzyme (two) 5. 11.8uL dH2O Used Pst1 + Spe1 for Lac. pr. and used Pst1 + Xba1 for tetR.
August 28 2007
RFP sub. transformation worked, but LacI did not. Picked colonies from RFP sub. and incubated in amp/LB overnight at 37 C. Supervisor informed that supercompotent cells used had been in a thaw accident, repeated LacI and RFP comp. transformations with new supercompotent cells using the same protocol
August 29 2007
RFP comp. worked (1 colony) but LacI once again did not. picked RFP comp. incubated in amp/LB overnight at 37 C.
Since LacI is not working, we are switching the repressor component of the reporter system. Lambda cI and a lambda cI inhibited promoter will be used instead of the LacI components.
Attempted Transformation of 3 Biobricks using XL1-blue supercompotent cells with same protocol as 27/08/07.
1. BBa_R0051 - Plate 1 well 9C - lambda cI inhibited promoter 2. BBa_C0051 - Plate 1 well 5G - lambda cI 3. BBa_J23119 - PLate 3 well 19A - Strong promoter for riboswitch
August 30 2007
All three transformations worked, picked two colonies from each and incubated overnight at 37 C in LB/amp
The RFP comp. colony picked on the 29th did not grow, but the plates had been left in the incubator for another night and colonies grew on both the LacI and RFP comp. plate. Probably false positives, but picked two colonies from each and inoculated LB/amp
September 3 2007
Created a new streak plate of RP1616 (cheZ mutant) to use with Fermentas TransformAid Kit to make cells competant for transformation.
Riboswitch PCR 37.5uL HF Buffer 7.5uL dNTP 80.7uL H2O 1.89uL Phusion 7.5uL Forward Primer 7.5uL Reverse Primer 2.5uL Template/Reaction --> 3 Reactions
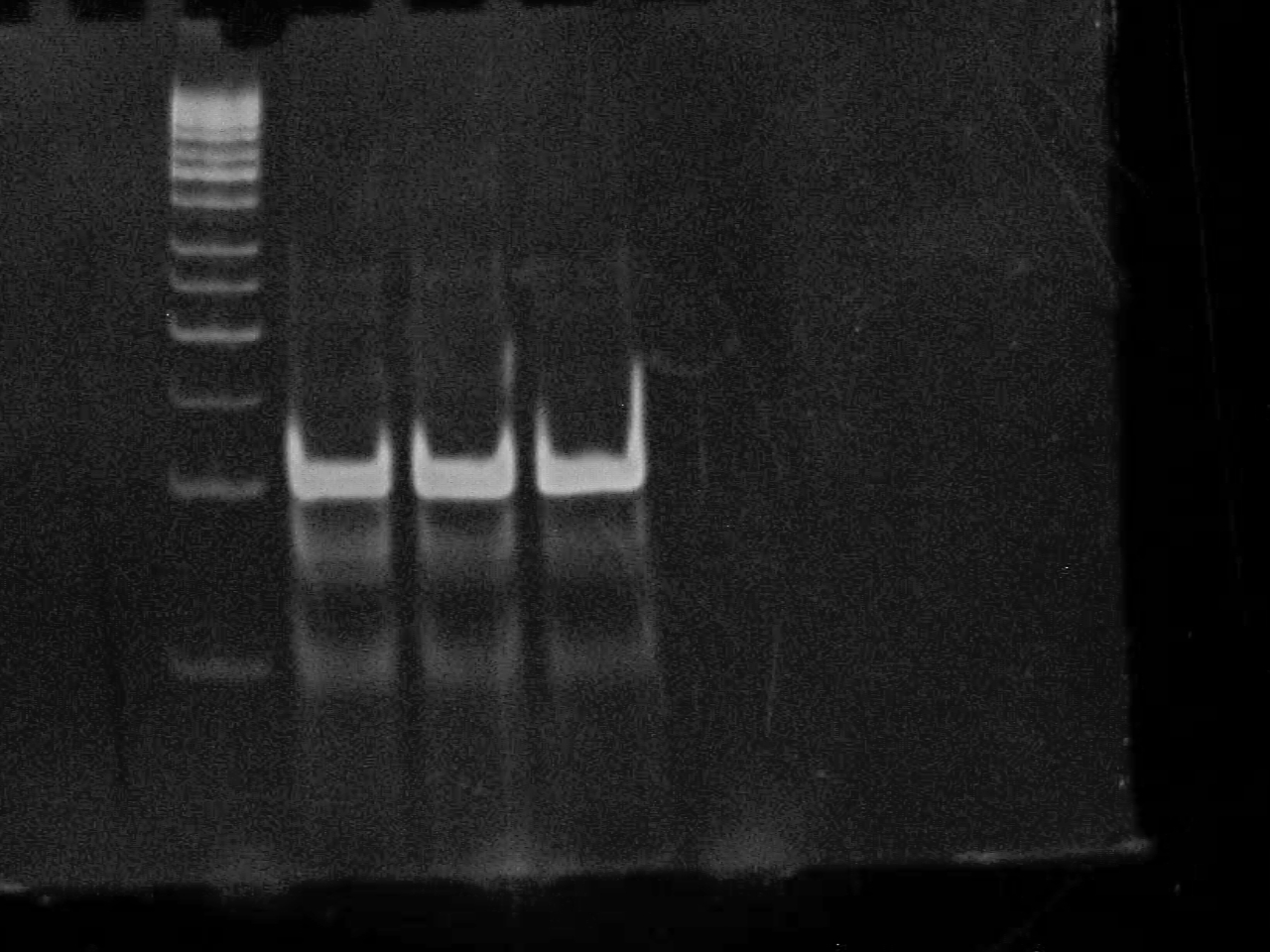
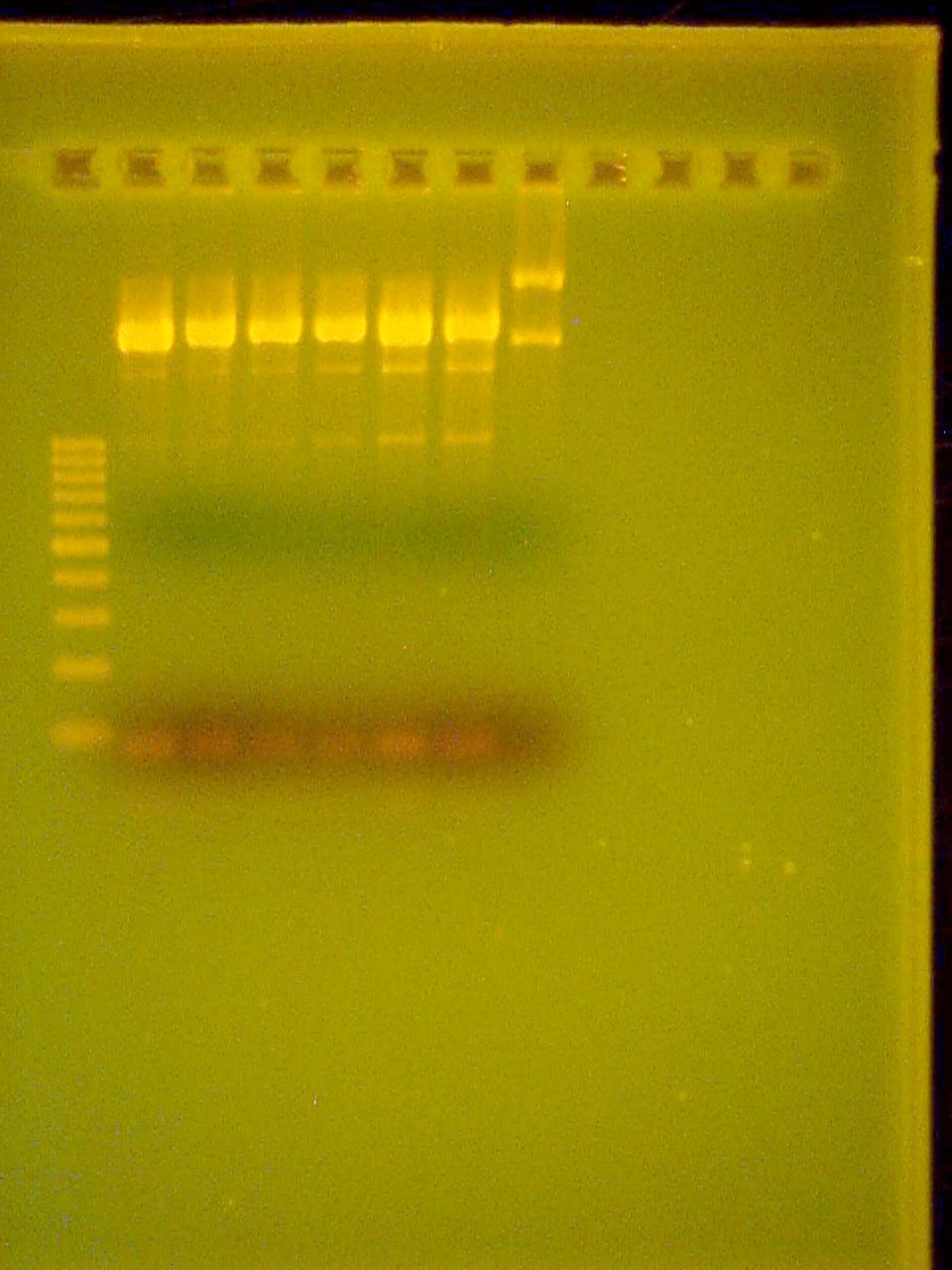
September 5 2007
Performed pruification of the PCR product of CheZ using Qiagen MinElute Kit for PCR purification.
Started overnight culture of RP1616 to use for transformation of Promoter+Riboswitch+CheZ plasmid as well as a puc19 control.
10% DNA PAGE Gel of Riboswitch PCR All three are positive!
September 5 2007
PCR Purification of Riboswitch using Qiagen MinElute Kit for PCR Purification
September 12 2007
Ran a digest of C0051, B0010, B0012, I13522, R0051, and I13507.
Reaction conditions 1. 5uL NEB 2 4. 1uL RE #1 2. 0.5uL BSA 5. 1uL RE #2 3. 5uL template 6. 37.5uL ddH2O Temp. 37C Overnight Enzymes used C0051 - EcoR1 + Spe1 I13522 - Xba1 + Pst1 B0010 - EcoR1 + Xba1 R0051 - Spe1 + Pst1 B0012 - Spe1 + Pst1 I13507 - Xba1 + Pst1
September 17 2007
[Riboswitch]=200ng/uL after purification
Double digestion of Riboswitch 5uL DNA 0.3uL EcoRI 0.3uL PstI 1uL NEBuffer 2 2.4 uL H2o 10uL Reaction - should be ~100ng/uL. Will try a sticky end and a blunt end ligation in parallel.'
September 18 2007
[pUC19] cut with EcorI and PstI is about 40ng/uL. Will use 10uL (400ng) for ligation. [pUC19] cut with SmaI is about 10ng/uL. Will use 10uL (100ng) for ligation.
Riboswitch ligations into pUC19
Sticky end 10uL pUC19 (400ng) 0.9uL riboswitch 2uL ligation buffer 2uL ligase (1U/uL) 5.1uL H2O
Blunt End 10uL pUC19 (100ng) 0.3uL riboswitch 2uL ligation buffer 1uL ligase 0.3uL SmaI 2uL PEG 4.4uL H2O
September 19 2007
PCR purification of newly made riboswitch
Transformation of Riboswitch in pUC19 into Ecoli DH5alpha. Plates contained XGal
Quantification gel of Sept 12 digests.
September 21 2007
Colony screening PCR of Riboswitch/pUC19
No. 1-8 - blunt end ligations No. 9-15 - sticky end ligations No. 16 and 17 - blue colonies No. 18 - PCR control (positive)
36uL 10x buffer 7.2uL dNTP 152.64uL H2O 2.16 uL Taq 18uL Forward primer (riboswitch primer) 18uL Reverse primer 36uL MgSO4
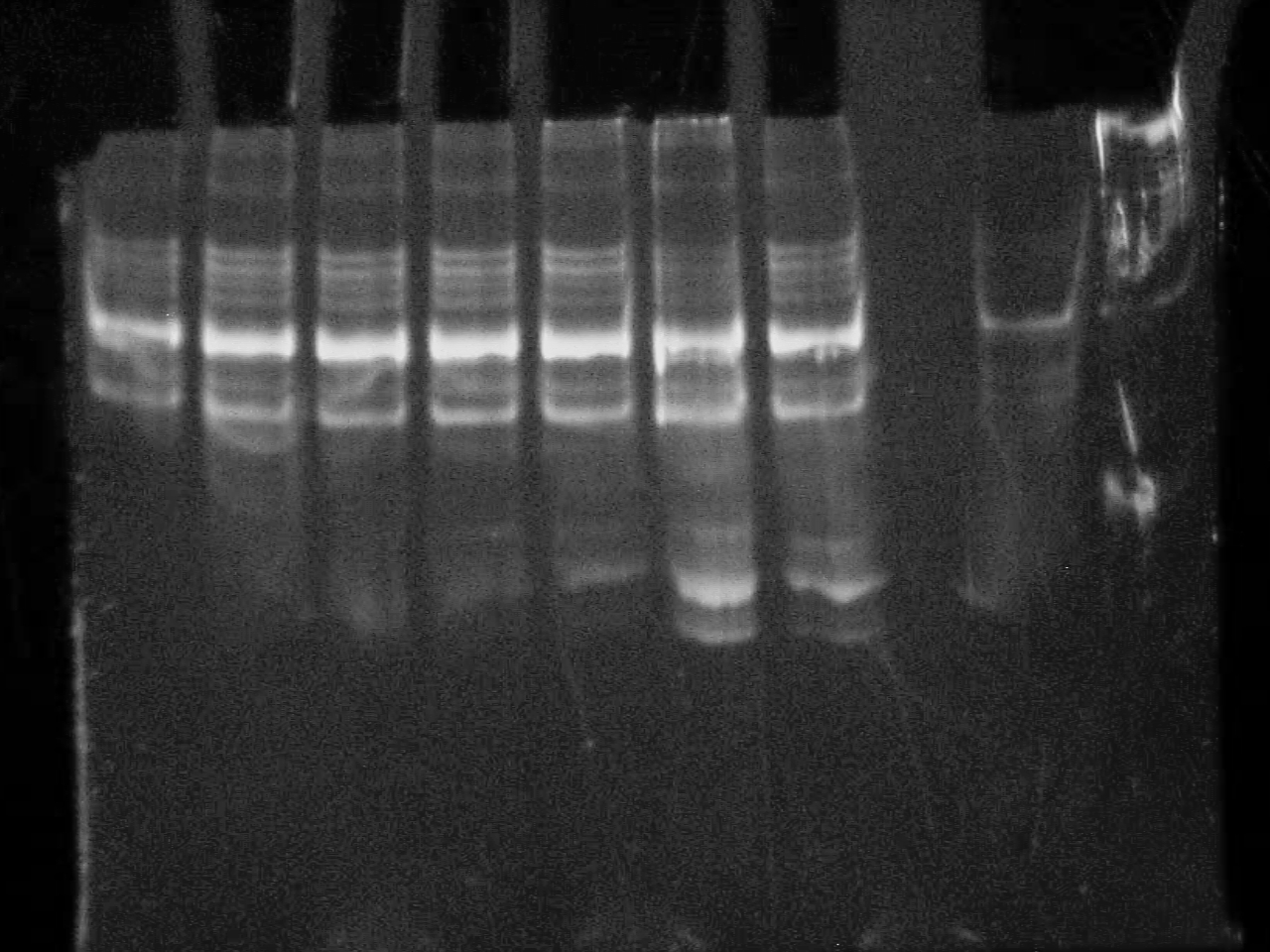
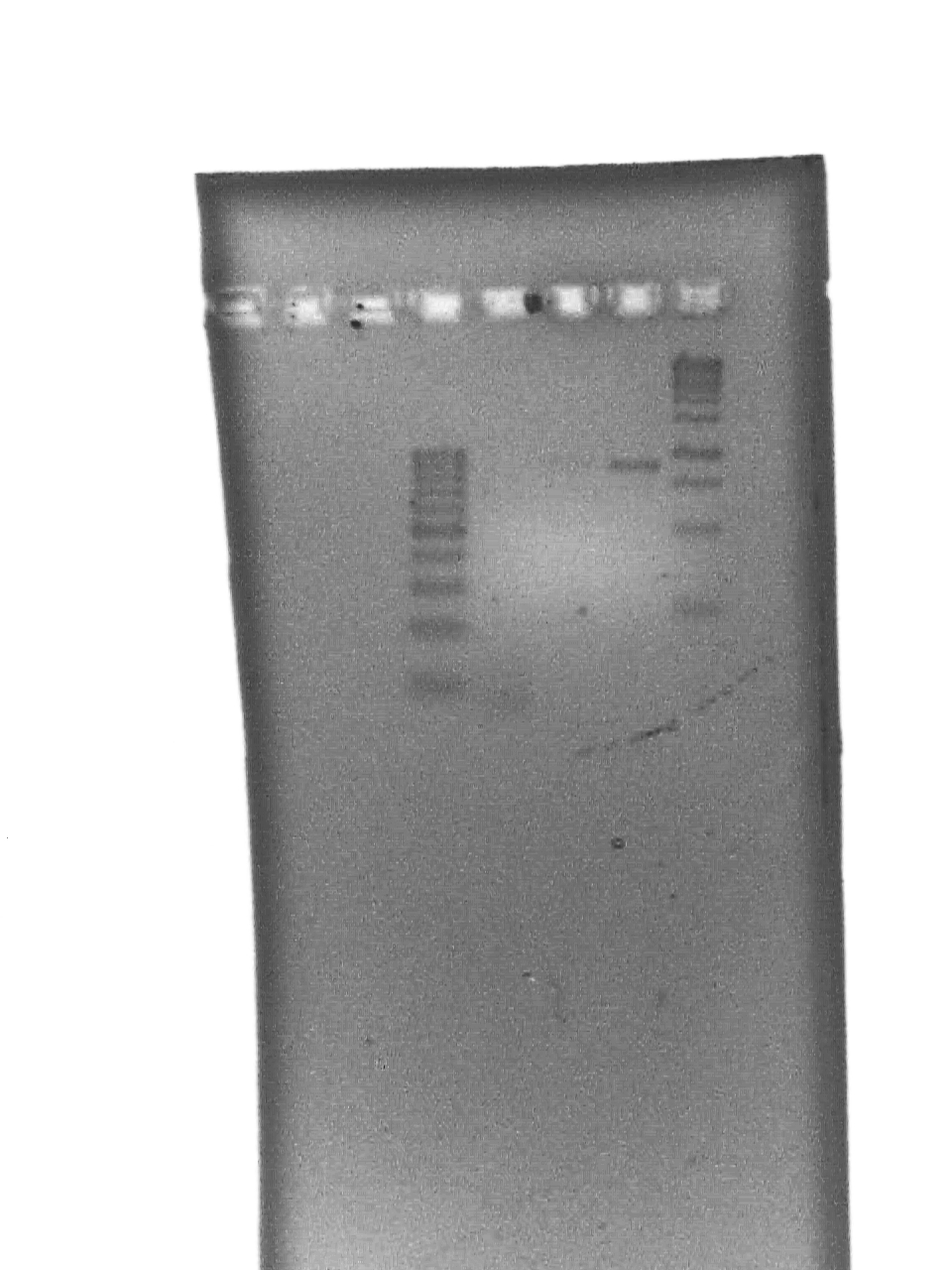
September 23 2007
10% DNA PAGE Gels of Colony screening.
All seem positive - will do a restriction with an enzyme that only cuts in the riboswitch
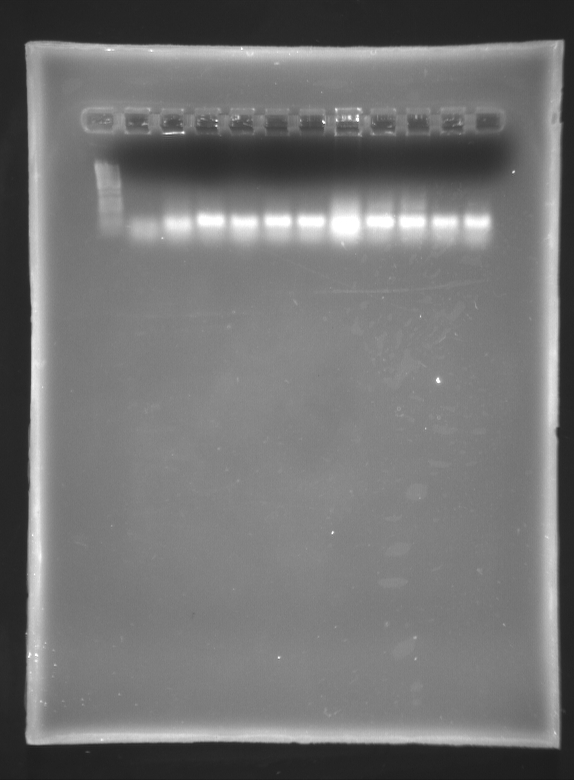
September 30 2007
Miniprepped Blunt No's 1,4 and 5, and Stick No's 9, 12, 15 using the QiaQuick spin Kit
Single restriction with SpeI
Mix 12uL NEBuffer 2 1.2 uL SpeI (2U) 1.2uL BSA 75.6uL H2O
5uL DNA per reaction, 6 reactions.
1% Agarose Gels - loaded 5uL of each sample
Number 1 1- 1Kb LAdder (4uL) 2- riboswitch 1 Unrestricted 3- " Restricted 4- riboswitch 4 Unrestricted 5- " Restricted 6- riboswitch 5 Unrestricted 7- " REstricted 8- riboswitch 9 Unrestricted 9- " Restricted 10- riboswitch 12 - Unrestricted 11 " Restricted 12 - empty
Number 2 1- riboswitch 15- unrestricted 2- " restricted 3-6 are other things 7- 1Kb ladder
October 20 2007
Double digestion of riboswitch in pUC19 with XbaI and PstI
Master Mix 8uL PstI 8uL XbaI 8uL NEBuffer II 4uL BSA 12uL H2O 5uL DNA in each
October 21 2007
Prepared semisolid plates containing theophylline or caffeine.
Plates were made with LB and 0.25% agar and ampicillin. 0mM, 0.1mM, 1mM, and 5mM Theophylline and Caffeine plates were made by adding the appropriate amount of 100mM stock solutions to 25mL of warm semisolid media before pouring plate.
A tube of DH5a cells containing the plasmid with CheZ and the riboswitch was grown overnight in LB with ampicillin.
Samples 1-6 seem to work. Pooled samples 5 and 6 for gel extration.
Preperative restriction 40uL DNA 2uL XbaI 2uL PstI 8uL NEBufferII 27uL H2O 1uL BSA
80uL total, incubate at 37C for two hours.
Gel extraction using Quaigen kit
2% Agarose gel to quantify:
Lane 1- 100bp ladder (4uL) Lane 2 - gel extracted riboswitch (4uL)
Concentration appears to be approximately 3.3 ng/uL
Ligation of Riboswitch into strong promoter plasmid. 3uL vector 4uL riboswitch insert 1uL ligase 1uL T4 DNA ligase buffer 1uL H2O Total of 10 uL
October 22 2007
Transformation of riboswitch/strong promotor plasmid
The plates from October 21 2007 were streaked with an inoculating loop to asses motility when cells were exposed various levels of theophylline and caffeine.
October 23 2007
Streaks made on October 22 2007 were assessed and the results were not clear. The variability present in initial streak size was larger than any difference in cell migration rate due to motility. On the highest concentration of Theophylline plated, there were a much larger number of satellite colonies formed than any other plates. This may be an indication of motility, but is far from conclusive.
Using the same plates, two drops (1uL and 10uL) of liquid culture were placed, with care to reduce variation in size. Plates were incubated overnight at 37 C.
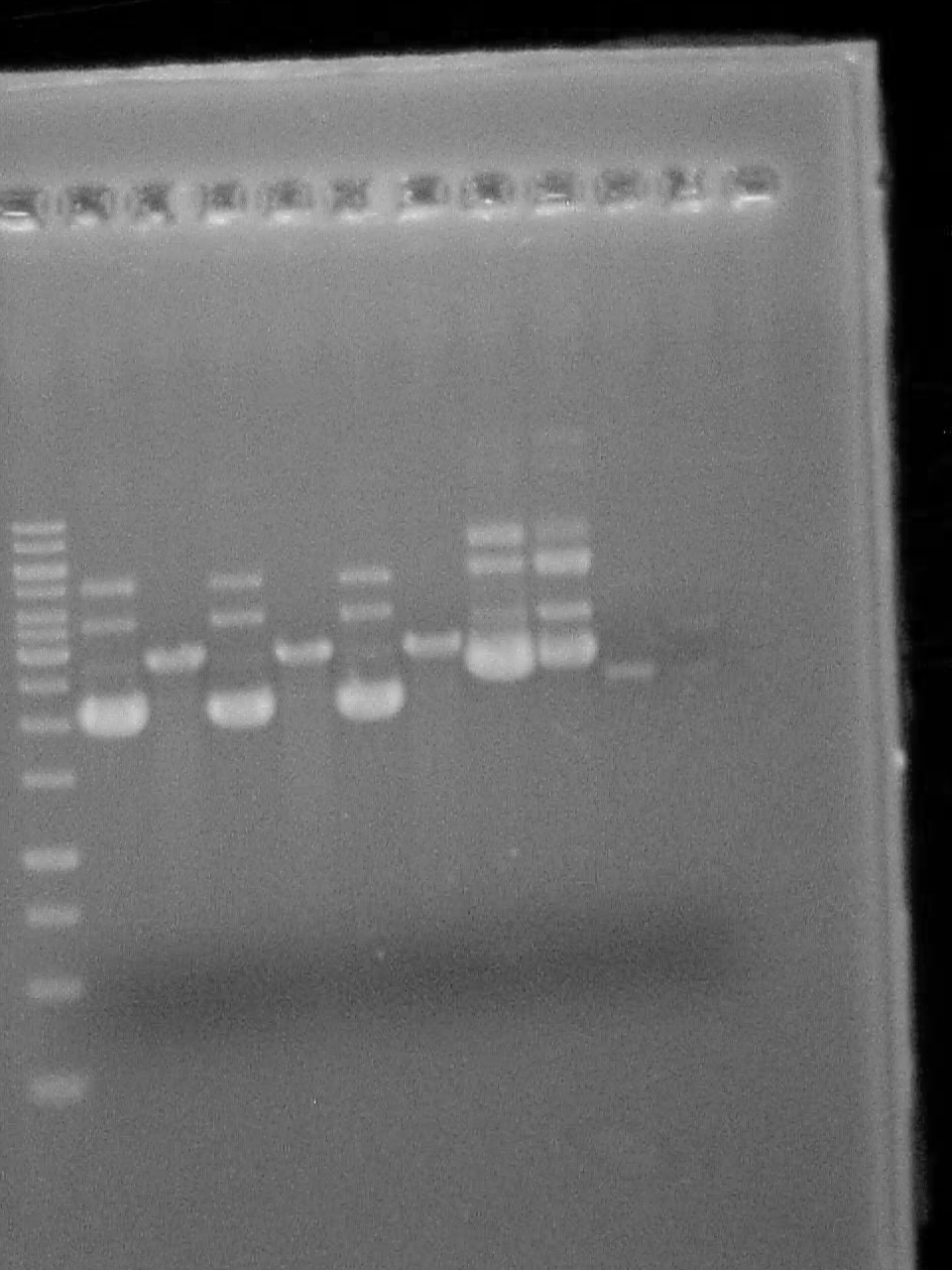
Picked 20 colonies for PCR colony screening
Master mix 25uL Forward primer 25uL Reverse primer 10uL dNTP 10X buffer 50uL 15.1uL H2O in each Swirl colony in each
Miniprepped colonies 5-10.
October 24 2007
The drops from October 23 2007 were assessed and no variation in size (presumably due to motility) was noticed above the variation present in drop size. This suggests that motility assays will need to be carried out at the microscopic level.
October 26 2007
Sent 2 pUC19 plasmids containing the riboswitch and CheZ as well as a pUC19 derivative plasmid containing a strong promoter with the riboswitch.
Main Page | Project Overview | Timeline | Lab Notebook | Team Members