NYMU Taipei/Lab Notes/2007 9 1
From 2007.igem.org
< NYMU Taipei/Lab Notes(Difference between revisions)
Lihsiangyen (Talk | contribs) |
|||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
*Plasmid extraction: RBS+cinR 1, RBS+HSL 1, RBS+HSL 2, RBS+HSL 3 from 5ml LB culture to 50ul EB | *Plasmid extraction: RBS+cinR 1, RBS+HSL 1, RBS+HSL 2, RBS+HSL 3 from 5ml LB culture to 50ul EB | ||
*The concentration was measured with spectrophotometer at 260nm | *The concentration was measured with spectrophotometer at 260nm | ||
| - | **RBS+cinR 1: | + | **RBS+cinR 1: 0.08ug/ul |
| - | **RBS+HSL 1: | + | **RBS+HSL 1: 0.03ug/ul |
| - | **RBS+HSL 2: | + | **RBS+HSL 2: 0.08ug/ul |
| - | **RBS+HSL 3: | + | **RBS+HSL 3: 0.06ug/ul |
*Enzyme digest RBS+cinR 1 | *Enzyme digest RBS+cinR 1 | ||
**RBS+cinR 1: 14ul | **RBS+cinR 1: 14ul | ||
| Line 43: | Line 43: | ||
*Comment | *Comment | ||
**There seems to be some incomplete digestion of RBS+HSL 2, possibly result from too many DNA. | **There seems to be some incomplete digestion of RBS+HSL 2, possibly result from too many DNA. | ||
| - | ** | + | **BsaBI digestion test of RBS+cinR failed. The lower bright band is probably negative supercoil form of the plasmid. Redo ligation RBS+cinR ver. 2. |
| + | **Assuming that the restriction enzyme is working, it should digest 1ug of DNA in 5min according to the NEB catalog. | ||
| + | **Todo: BsaBI digestion of cinR | ||
| + | *Ligation (RBS+cinR+RBS+HSL) | ||
| + | **RBS+cinR: 1ul | ||
| + | **RBS+HSL 3: 3ul | ||
| + | **10x buffer A: 1ul | ||
| + | **10x ligase (Yeastern, 2U/ul): 1ul | ||
| + | **H2O: 4ul | ||
| + | **Incubated at RT for 20min | ||
| + | *Ligation (RBS+cinR ver. 2) | ||
| + | **RBS: 1ul | ||
| + | **cinR: 1ul | ||
| + | **10x buffer A: 1ul | ||
| + | **10x ligase (Yeastern, 2U/ul): 1ul | ||
| + | **H2O: 6ul | ||
| + | **Incubated at RT for 20min | ||
Latest revision as of 06:23, 16 September 2007
- Plasmid extraction: RBS+cinR 1, RBS+HSL 1, RBS+HSL 2, RBS+HSL 3 from 5ml LB culture to 50ul EB
- The concentration was measured with spectrophotometer at 260nm
- RBS+cinR 1: 0.08ug/ul
- RBS+HSL 1: 0.03ug/ul
- RBS+HSL 2: 0.08ug/ul
- RBS+HSL 3: 0.06ug/ul
- Enzyme digest RBS+cinR 1
- RBS+cinR 1: 14ul
- SpeI: 1ul
- PstI 1ul
- 10x buffer 2: 2ul
- 10x BSA: 2ul
- 37°C water bath, 2h
- Heat inactivation: 65°C, 20min
- Enzyme digest RBS+HSL 2
- RBS+HSL 2: 14ul
- XbaI: 1ul
- PstI: 1ul
- 10x buffer 3: 2ul
- 10x BSA: 2ul
- 37°C water bath, 2h
- Heat inactivation: 65°C, 20min
- Enzyme digest RBS+HSL 3
- RBS+HSL 3: 14ul
- XbaI: 1ul
- PstI: 1ul
- 10x buffer 3: 2ul
- 10x BSA: 2ul
- 37°C water bath, 2h
- Heat inactivation: 65°C, 20min
- Check RBS+cinR 1 with BsaBI digestion
- RBS+cinR 1: 3ul
- BsaBI: 1ul
- 10x buffer 2: 2ul
- H2O: 14ul
- 60°C dry bath, 1h
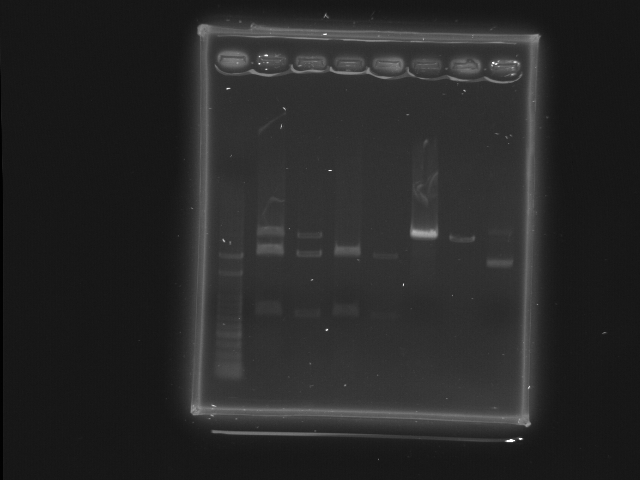
- Well 1: 100pb ladder
- Well 2 3: "RBS+HSL 2" XbaI, PstI
- Well 4 5: "RBS+HSL 3" XbaI, PstI
- Well 6 7: "RBS+cinR 1" SpeI, PstI
- Well 8: "RBS+cinR 1" BsaBI
- Comment
- There seems to be some incomplete digestion of RBS+HSL 2, possibly result from too many DNA.
- BsaBI digestion test of RBS+cinR failed. The lower bright band is probably negative supercoil form of the plasmid. Redo ligation RBS+cinR ver. 2.
- Assuming that the restriction enzyme is working, it should digest 1ug of DNA in 5min according to the NEB catalog.
- Todo: BsaBI digestion of cinR
- Ligation (RBS+cinR+RBS+HSL)
- RBS+cinR: 1ul
- RBS+HSL 3: 3ul
- 10x buffer A: 1ul
- 10x ligase (Yeastern, 2U/ul): 1ul
- H2O: 4ul
- Incubated at RT for 20min
- Ligation (RBS+cinR ver. 2)
- RBS: 1ul
- cinR: 1ul
- 10x buffer A: 1ul
- 10x ligase (Yeastern, 2U/ul): 1ul
- H2O: 6ul
- Incubated at RT for 20min