Edinburgh/DivisionPopper/Design
From 2007.igem.org
Luca.Gerosa (Talk | contribs) |
|||
| (118 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | '''MENU''' :[[Edinburgh/DivisionPopper| Introduction]] | [[Edinburgh/DivisionPopper/References|Background]] | [[Edinburgh/DivisionPopper/Applications|Applications]] | [[Edinburgh/DivisionPopper/Design|Design]] | | + | '''MENU''' :[[Edinburgh/DivisionPopper| Introduction]] | [[Edinburgh/DivisionPopper/References|Background]] | [[Edinburgh/DivisionPopper/Applications|Applications]] | [[Edinburgh/DivisionPopper/Design|Design&Implementation]] | [[Edinburgh/DivisionPopper/Modelling|Modelling]] | [[Edinburgh/DivisionPopper/Status|Wet Lab]] | [[Edinburgh/DivisionPopper/SBApproach|Synthetic Biology Approach]] | [[Edinburgh/DivisionPopper/Conclusions|Conclusions]] |
| - | In the Design section we define the architecture ( | + | In the Design section we define the architecture (design), the biology elements (implementation) and the mechanisms (dynamics) of the device. According to the [http://partsregistry.org/cgi/htdocs/AbstractionHierarchy/index.cgi abstraction levels hierarchy], we propose a view of the Division PoPper at the level of device, parts and DNA. Regarding to the dynamics, we explain which biological processes are involved and how they cooperate in defining the behaviour of the device. The logic design and implementation phases have been decoupled. |
__TOC__ | __TOC__ | ||
| - | |||
| - | |||
| - | == | + | ==Design== |
| - | + | In this section we explain the functional components of the device and how they are connected logically and physically. This view of the device follows the [http://partsregistry.org/cgi/htdocs/AbstractionHierarchy/index.cgi abstraction hierarchy] proposed in the Registry of Parts. | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | [[Image: | + | {| width="20%" align="right" style="text-align:center" |
| + | |- | ||
| + | |'''Device abstraction level of the Division Counter''' | ||
| + | |- | ||
| + | |[[Image:Devicelevel.png|350px]] The Division PoPper device generates an input in the form of PoPS signal. | ||
| + | |} | ||
| - | |||
| - | |||
| - | === | + | ===Device abstraction level === |
| - | + | The Division PoPper is a device, because it is a functional element that can be further combined with other devices by the use of a standard PoPS signal. The device has no formal input but its behaviour is triggered by an "external" stimulus: the cell division. | |
| + | The output is a PoPS signal that is supposed to be always zero (not considering leaking issues in the promoters) apart from the short period immediately subsequent to a cell division event. In this case the device generates a pulse: the PoPS signal reaches a non-zero level for a period of time, returning to zero afterward. The function of the device is simple: to inform a downstream device that a division has happened by sending a pulse signal. We suggest some system configurations in the [[Edinburgh/DivisionPopper/Applications|Applications]] section. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | {| width="20%" align="right" style="text-align:center" | |
| + | |- | ||
| + | |'''Part abstraction level (Design)''' | ||
| + | |- | ||
| + | |[[Image:Partslevel(logic).png|350px]] Design of the part structure in the Division PoPper device. | ||
| + | |} | ||
| - | + | ===Part abstraction level === | |
| - | |||
| - | |||
| - | + | At the level of part abstraction we define the functional parts of the device: regulatory regions, coding regions, terminators and so on. Instead of proposing directly the actual biological entities, we propose here a logic view. Strictly speaking, we define entities by their attributes and abilities, but without mapping them to real biological objects (his is done in the Implementation section below). The motivation is to separate the functional design from the realization choices. | |
| - | + | '''The Division PoPper functional parts are:''' | |
| - | + | * two recombinase site sequences: Rec. | |
| + | * a promoter A that can be repressed by protein A: promoter A. | ||
| + | * a coding region for protein A: repressor A. | ||
| + | * a promoter B that can be repressed by protein B: promoter B. | ||
| + | * a coding region for protein B: repressor B. | ||
| + | * a promoter P that is constitutively active (expressing). | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | '''The functional ability that these entities should have are:''' | |
| - | + | * the region between Rec recombinase site sequence is inverted at each cell division. | |
| + | * the protein A and protein B degradation rates should be fast enough to degrade them rapidly when not expressed. | ||
| + | * the protein A and protein B effects on (respectively) the promoter A and B expression capacity should be strong. | ||
| + | * promoter P should be strong in its constitutive expression. | ||
| - | |||
| - | ''' | + | '''The physical configuration:''' |
| - | + | The design relies on the alternate expression of the two strands of the DNA segment in between the Recombinase sites. Promoter P is placed outside the "flipping region" and controls its downstream region in the upper strand. The output of the device is given by the PoPS activity on the upper strand downstream from the second recombinase site. Since the region in between the recombinase sites flips, the output is controlled alternatively by the two DNA strands. In one strand the parts sequence is: repressor A, terminator, Promoter A. In the other the sequence is: repressor B, terminator, Promoter B. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | + | '''The dynamics:''' | |
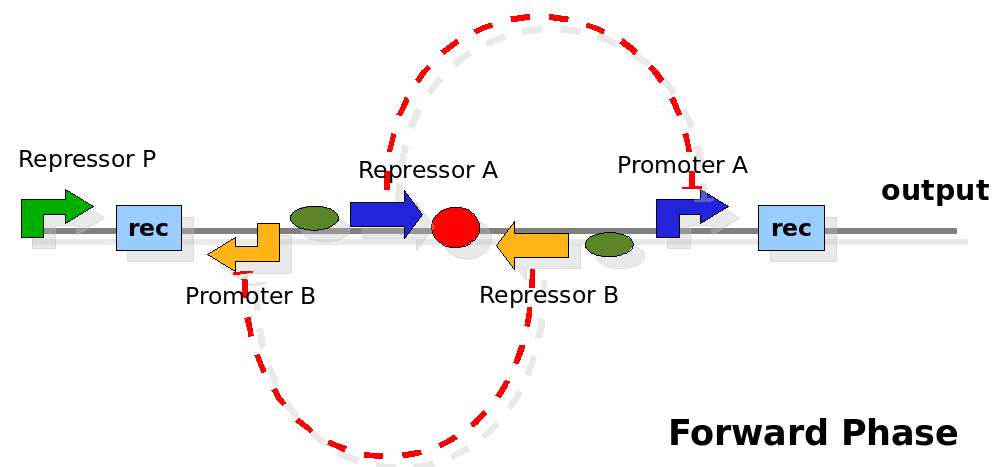
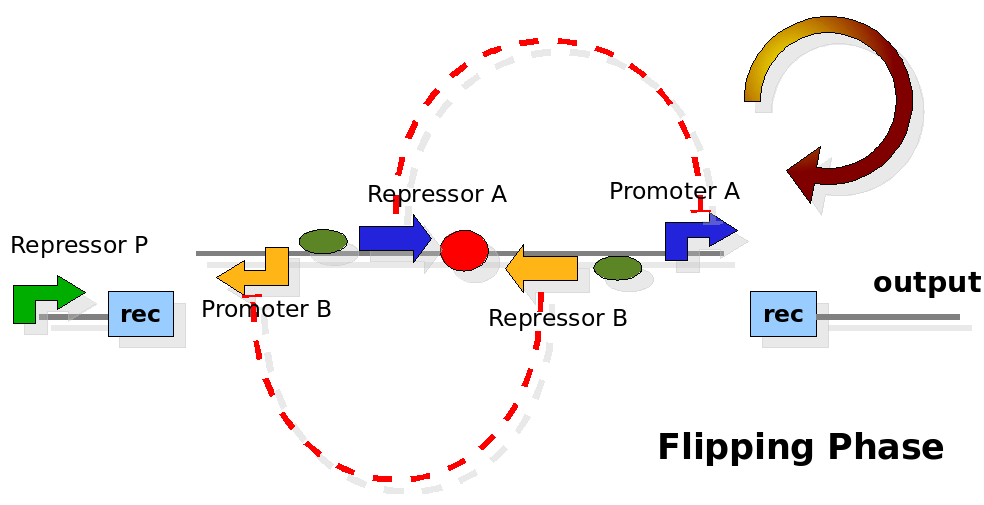
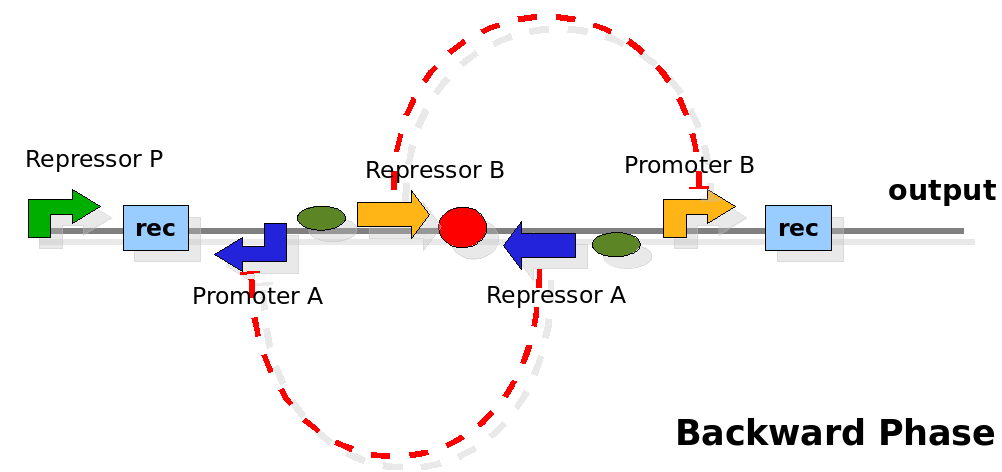
| - | {| | + | As cell division occurs, DNA enclosed by two inverted Recombinase sites is inverted. Initially the device is in the configuration with Repressor A and Promoter A on the upper strand. There is no Repressor A present in the cell, so Promoter A produces a constant PoPS output. As time passes, Repressor A accumulates and 'turns off' Promoter B. Meanwhile Repressor B is no produced and degrades so that, at the next division, Promoter B can produce PoPS output and the process repeats. This mechanism can be seen as the sequence of three phases: Forward, Flipping and Backward. We explain for each phases the dynamics: |
| - | ! | + | |
| + | {| border="1" cellpadding="20" cellspacing="0" | ||
| + | !Configuration | ||
| + | !Processes | ||
| + | !Output | ||
|- | |- | ||
| - | | | + | |[[Image:PoPperforward.png|350px]] |
| + | | During this phase, the output of the device is controlled by Promoter A. We assume there is no Repressor A present initially. Thus Promoter A generates a flux of PoPS output. After a certain amount of time, Repressor A builds up, since the constitutive promoter P placed upstream controls is expression. When the concentration of Repressor A is high enough, the flux of PoPS from the output promoter goes to zero. Meanwhile, the expression of Repressor B is not active. Thus the concentration of Repressor B should decrease to a low level. | ||
| + | | Controlled by Promoter A. | ||
|- | |- | ||
| - | + | |[[Image:PoPperflipping.png|350px]] | |
| + | | When a division of the cell happens, the recombinase system is activated. During this phase there is no physical active connection between the output region and a promoter, thus the PoPS flux is considered to be zero. | ||
| + | | Not controlled, expression should be zero. | ||
|- | |- | ||
| - | | | + | |[[Image:PoPperbackward.png|350px]] |
| + | | During this phase, the output of the device is controlled by promoter B. We assume there is a low Repressor B concentration at this time. Thus Promoter B generates a flux of PoPS output. After a certain amount of time, Repressor B accumulates, since the constitutive promoter P placed upstream controls its expression. When the concentration of Repressor B is sufficient, the flux of PoPS goes to zero. Meanwhile, the expression of Repressor A is not active. Thus the concentration of Repressor A should decrease to a low level. | ||
| + | |Controlled by Promoter B. | ||
|- | |- | ||
| - | | | + | |} |
| + | |||
| + | ==Implementation== | ||
| + | |||
| + | In the previous section we define which functional components are needed at the level of parts. In this section the members with biology competences evaluate the design and identify the actual biological parts that can be used. Further on, the DNA abstraction level details are proposed. | ||
| + | |||
| + | {| width="20%" align="right" style="text-align:center" | ||
|- | |- | ||
| - | | | + | |'''Part abstraction level (Implementation)''' |
|- | |- | ||
| - | | | + | |[[Image:partslevel.png|350px]] Implementation of the part structure in the Division PoPper device. |
|} | |} | ||
| - | |||
| - | + | ===Part abstraction level=== | |
| - | ! | + | |
| + | '''The Division PoPper parts are:''' | ||
| + | |||
| + | * two Dif recombinase sites (design: Rec). | ||
| + | * Tet promoter (design: promoter A). | ||
| + | * TetR repressor (design: repressor A). | ||
| + | * CLambda promoter (design: promoter B). | ||
| + | * CLambda repressor (design: repressor B). | ||
| + | * Lac promoter (design: promoter P). | ||
| + | |||
| + | The Registry of Standard Parts contains already some of the elements we need, for the others we constructed and added their BioBricked version. | ||
| + | |||
| + | '''The Division PoPper parts registry entries are:''' | ||
| + | |||
| + | * two Dif recombinase sites: [http://partsregistry.org/Part:BBa_I742101 BBa_I742101],[http://partsregistry.org/Part:BBa_I742102 BBa_I742102] | ||
| + | * Tet promoter:[http://partsregistry.org/Part:BBa_R0040 BBa_R0040] | ||
| + | * TetR repressor:[http://partsregistry.org/Part:BBa_C0040 BBa_C0040] | ||
| + | * CLambda promoter:[http://partsregistry.org/Part:BBa_R0051 BBa_R0051] | ||
| + | * CLambda repressor:[http://partsregistry.org/Part:BBa_C0051 BBa_C0051] | ||
| + | * Lac promoter:[http://partsregistry.org/Part:BBa_I14032 BBa_I14032] | ||
| + | |||
| + | Click on the links for details. | ||
| + | |||
| + | |||
| + | '''The assumptions regarding the biological elements are:''' | ||
| + | |||
| + | * The Dif sites flip only once during each cell division | ||
| + | * TetR and ClambdaR are produced and degrade faster than the cell cycle | ||
| + | |||
| + | For this device to work, ''dif'' site-enclosed DNA needs to flip only once per division. Like all recombination sites, ''dif'' sites are directional and bacteria use directly repeated ''dif'' sites to resolve genome dimers. Research shows that this resolution occurs only at septation, and we use this temporal control to inverse a plasmid-borne sequence to induce different functions. For more information look at the [[Edinburgh/DivisionPopper/References|Background]] section. | ||
| + | |||
| + | '''The Biology Processes:''' | ||
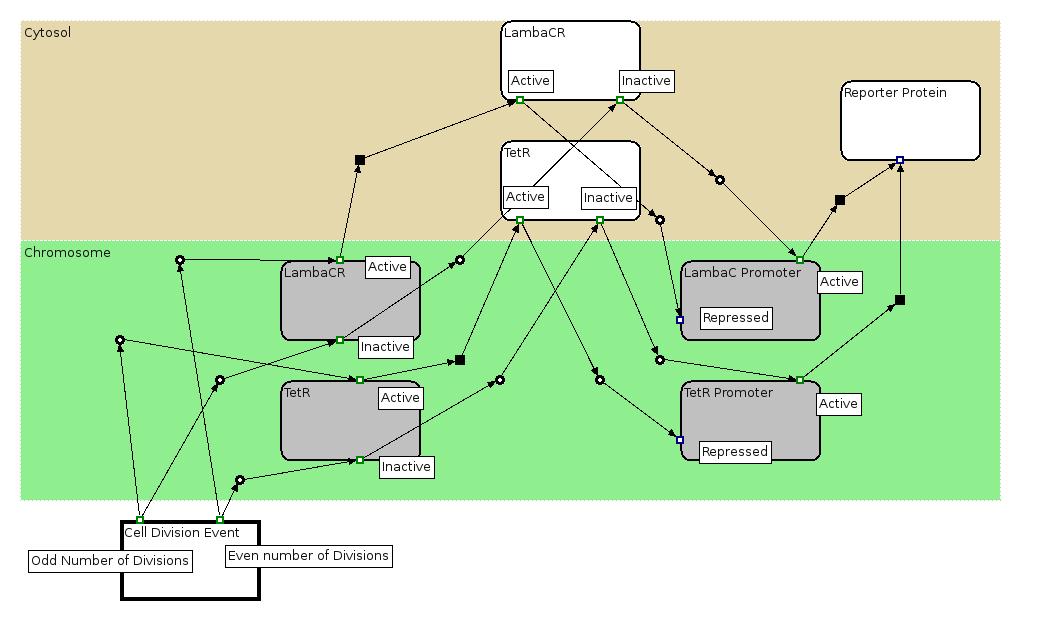
| + | We used the Edinburgh Pathway Notation, a state transition graphical notation to express the biological processes of the Division PoPper. The details regarding the notations can be found in the paper | ||
| + | [http://www-bm.ipk-gatersleben.de/stable/php/journal/articles/pdf/jib-36.pdf A Graphical Notation to Describe the Logical Interactions of Biological Pathways]. | ||
| + | |||
| + | |||
| + | {| border="1" cellpadding="20" cellspacing="0" | ||
| + | !Diagram | ||
| + | !Explanation | ||
|- | |- | ||
| - | | | + | |[[Image:PoPperEPN.jpg|550px]] |
| + | |This EPN Diagram explains which are the state configuration of the Division PoPper construct when a Reporter Protein is placed downstream the device. The Division Event state explains which state transitions occur when a division happens. The gray boxes represent the state (active, inactive) of the coding regions and promoter regions associated to TetR and LambaCR production. The white boxes represent the proteins. | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| width="20%" align="right" style="text-align:center" | ||
| + | |- | ||
| + | |'''DNA strands configuration''' | ||
| + | |- | ||
| + | |[[Image:DNAstrands.png|350px]] This graph explain on which DNA strand the parts are placed and with which coding direction. | ||
| + | |} | ||
| + | |||
| + | ===DNA abstraction level=== | ||
| + | |||
| + | In order for this construct to be implemented as BioBricks combined in the normal fashion, it was necessary for some of the components to be synthesised in inverse orientation. The final list of components for the sequence, in order, is as follows: | ||
| + | *''lac'' promoter [http://partsregistry.org/Part:BBa_I14032 BBa_I14032] | ||
| + | *''dif'' site in forward orienation, generated by us as [http://partsregistry.org/Part:BBa_I742101 BBa_I742101]. (physically generated as a BioBrick and deposited). | ||
| + | *reverse version of lambda cI-controlled promoter [http://partsregistry.org/Part:BBa_R0051 BBa_R0051]: designed as [http://partsregistry.org/Part:BBa_I742126 BBa_I742126]. | ||
| + | *ribosome binding site [http://partsregistry.org/Part:BBa_B0034 BBa_B0034]. | ||
| + | *Tet repressor coding sequence with rapid degradation tag [http://partsregistry.org/Part:BBa_C0040 BBa_C0040]. | ||
| + | *bidirectional terminator [http://partsregistry.org/Part:BBa_B0011 BBa_B0011] (initially it was planned to use a combination of several terminators here in different orientations, but after consultation with DNA synthesis company GeneArt, it was considered that the repeated sequences would cause difficulties in synthesis. If termination appears to be insufficient, sub-regions of the device can be extracted as separate biobricks by PCR, and further terminators inserted). In the final design submited to GeneArt, for certain reasons an inverse version of BBa_B0011 was used, and this was entered in our sandbox as [http://partsregistry.org/Part:BBa_I742129 BBa_I742129]. | ||
| + | *Reverse coding sequence of lambda cI repressor BBa_C0051, entered in our sandbox as [http://partsregistry.org/Part:BBa_I742131 BBa_I742131]. | ||
| + | *Reverse ribosome binding site BBa_B0034, entered in our sandbox as [http://partsregistry.org/Part:BBa_I742130 BBa_I742130]. Since the normal abbreviated biobrick scar between RBS and coding sequence could not be used in the inverse orientation, the length of the RBS was decreased by two bases to maintain the proper spacing. | ||
| + | *Tet repressor-controlled promoter [http://partsregistry.org/Part:BBa_R0040 BBa_R0040]. | ||
| + | *''dif'' site in reverse orientation, generated as [http://partsregistry.org/Part:BBa_I742102 BBa_I742102] and deposited. | ||
| + | *The total construct has been entered in our sandbox as [http://partsregistry.org/Part:BBa_I742132 BBa_I742132]. | ||
| + | |||
| + | In order for the device to be tested, it will be necessary to insert it into a '''single copy BioBrick vector''', since if multiple copies are present the likelihood is increased that not all will be in the same orientation, leading to simultaneous production of both repressors and lack of output (this cross-talk between separate instances of the device within a single system is a characteristic feature of biological systems which must always be considered) . For this purpose, the single copy F'-episome-based plasmid '''pHR277''' (kanR) was generously provided by Professor Millicent Masters, Institute of Cell Biology, University of Edinburgh. This plasmid possesses unique XbaI and PstI sites which can be used to insert a biobrick; unfortunately, it also possesses an EcoRI site elsewhere on the plasmid so, unless this is removed, it cannot be converted into a fully compliant vector which can be used to assemble new BioBricks (in contrast to our Gram positive BioBrick vector pTG262-BB (BBa_I742123), described elsewhere in this wiki). | ||
| + | |||
| + | ==Proofs of Concept== | ||
| + | |||
| + | The complex structure of the Division PoPper and the small amount of time we have for the construction of the device in the context of the iGEM competition led us to plan the construction of a simpler construct: the Proof of Concept device. The aim of the Proof of Concept device is to test the main assumptions at the basis of the Division PoPper device: that the recombinase mechanism is activated only once and at cell division and that the time between divisions is sufficient for the repression mechanism to be activated and inactivated. We planned to construct two experiments: | ||
| + | |||
| + | '''Experiment 1:''' | ||
| + | |||
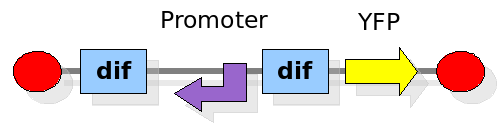
| + | [[Image:proof1backward.png|300px|float|right]] | ||
| + | |||
| + | This experiment will prove whether or not the DNA between the two ''dif'' sites flips at all during cell division. If the a flip occurs, then the direction the promoter operates will be changed and YFP will be expressed. The BioBrick used for YFP expression is [http://partsregistry.org/Part:BBa_E0430 BBa_E0430]. The reverse ''lac'' promoter has been constructed and deposited as [http://partsregistry.org/Part:BBa_I742124 BBa_I742124]. | ||
| + | |||
| + | |||
| + | '''Experiment 2:''' | ||
| + | |||
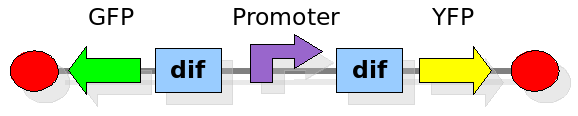
| + | [[Image:proof2forward.png|300px|float|right]] | ||
| + | |||
| + | This investigates the number of flips that occurs during division and will require rapidly degrading fluorescent proteins. | ||
| + | After each division we expect to see a change in colour as the promoter activates a different reporter. We developed some mathematical models of this scenario that can be seen in the [[Edinburgh/DivisionPopper/Modelling|Modelling]] section. In the [[Edinburgh/DivisionPopper/Status|Wet Lab]] section you can find all the detail regarding the construction in the lab. | ||
| + | |||
| + | |||
| + | {| border="1" cellpadding="20" cellspacing="0" | ||
| + | !Diagram | ||
| + | !Explanation | ||
|- | |- | ||
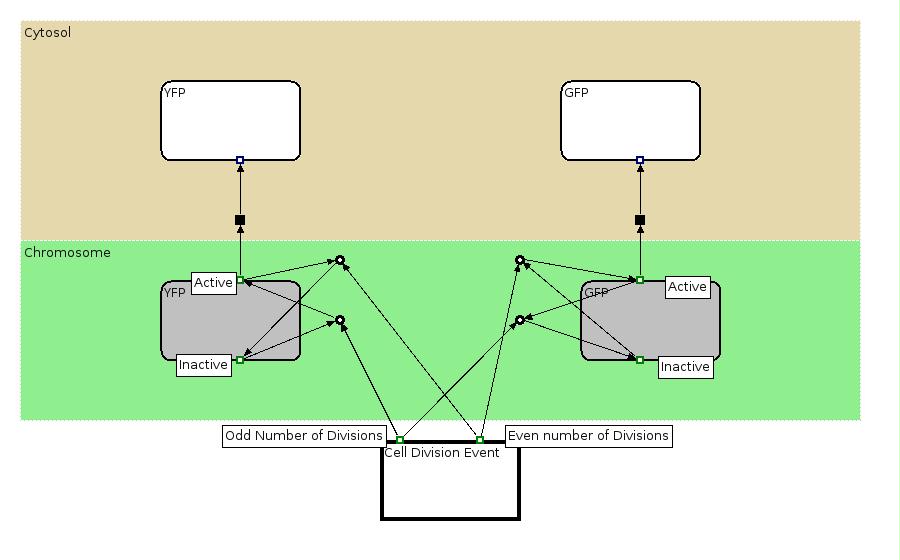
| - | | | + | |[[Image:Proof2EPN.jpg|550px]] |
| + | |This EPN Diagram explains which are the state configuration of the Proof of Concept construct with YFP and GFP production. The Division Event state explains which state transitions occur when a division happens. The grey boxes represent the state (active, inactive) of the genes associated to YFP and GFP production and the white boxes represent the two proteins. | ||
|- | |- | ||
| - | |||
|} | |} | ||
Latest revision as of 20:50, 26 October 2007
MENU : Introduction | Background | Applications | Design&Implementation | Modelling | Wet Lab | Synthetic Biology Approach | Conclusions
In the Design section we define the architecture (design), the biology elements (implementation) and the mechanisms (dynamics) of the device. According to the [http://partsregistry.org/cgi/htdocs/AbstractionHierarchy/index.cgi abstraction levels hierarchy], we propose a view of the Division PoPper at the level of device, parts and DNA. Regarding to the dynamics, we explain which biological processes are involved and how they cooperate in defining the behaviour of the device. The logic design and implementation phases have been decoupled.
Contents |
Design
In this section we explain the functional components of the device and how they are connected logically and physically. This view of the device follows the [http://partsregistry.org/cgi/htdocs/AbstractionHierarchy/index.cgi abstraction hierarchy] proposed in the Registry of Parts.
| Device abstraction level of the Division Counter |
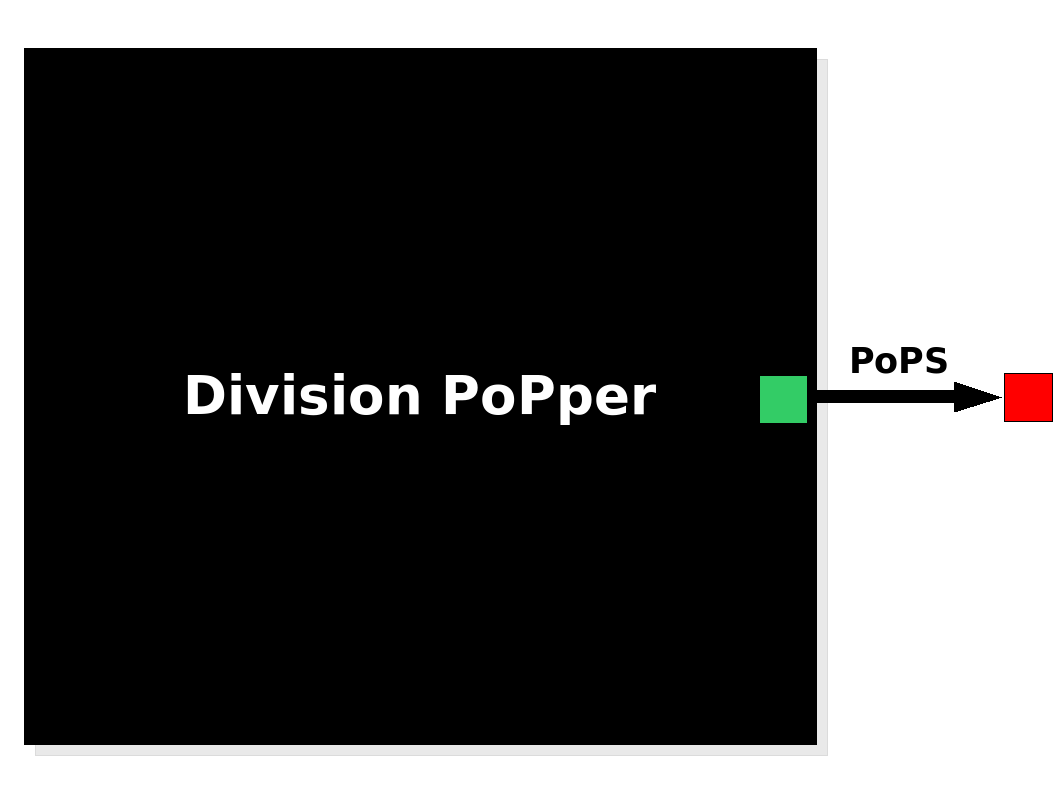 The Division PoPper device generates an input in the form of PoPS signal. The Division PoPper device generates an input in the form of PoPS signal.
|
Device abstraction level
The Division PoPper is a device, because it is a functional element that can be further combined with other devices by the use of a standard PoPS signal. The device has no formal input but its behaviour is triggered by an "external" stimulus: the cell division. The output is a PoPS signal that is supposed to be always zero (not considering leaking issues in the promoters) apart from the short period immediately subsequent to a cell division event. In this case the device generates a pulse: the PoPS signal reaches a non-zero level for a period of time, returning to zero afterward. The function of the device is simple: to inform a downstream device that a division has happened by sending a pulse signal. We suggest some system configurations in the Applications section.
| Part abstraction level (Design) |
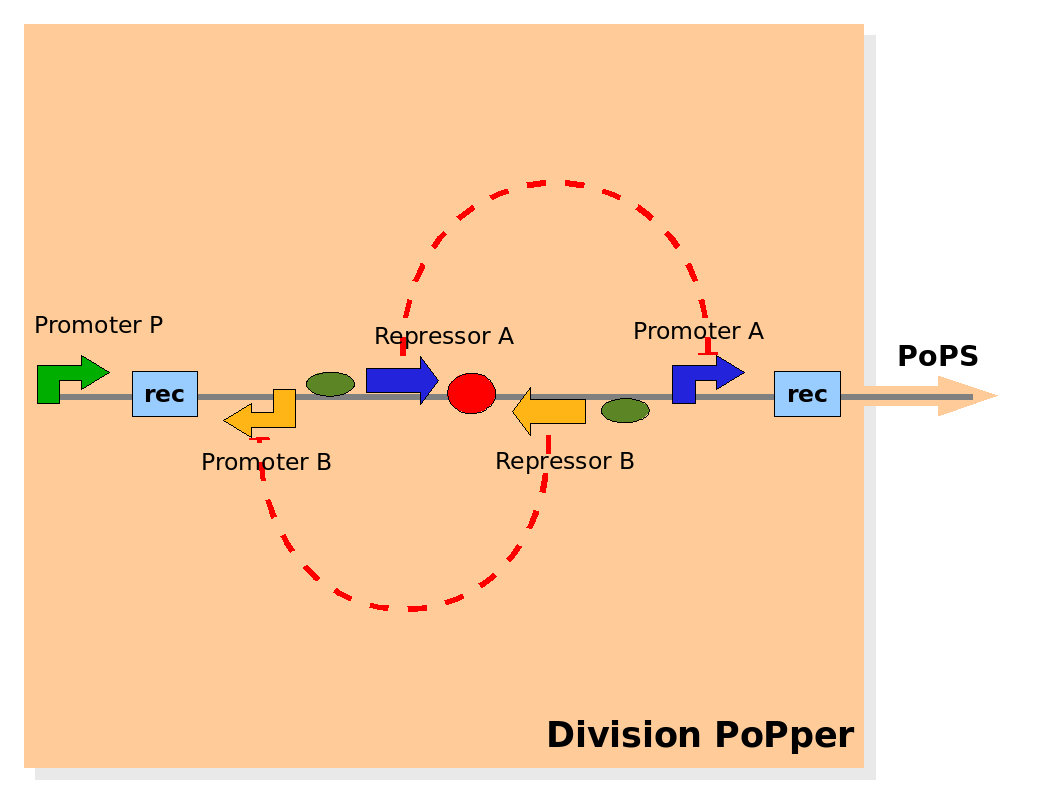 Design of the part structure in the Division PoPper device. Design of the part structure in the Division PoPper device.
|
Part abstraction level
At the level of part abstraction we define the functional parts of the device: regulatory regions, coding regions, terminators and so on. Instead of proposing directly the actual biological entities, we propose here a logic view. Strictly speaking, we define entities by their attributes and abilities, but without mapping them to real biological objects (his is done in the Implementation section below). The motivation is to separate the functional design from the realization choices.
The Division PoPper functional parts are:
- two recombinase site sequences: Rec.
- a promoter A that can be repressed by protein A: promoter A.
- a coding region for protein A: repressor A.
- a promoter B that can be repressed by protein B: promoter B.
- a coding region for protein B: repressor B.
- a promoter P that is constitutively active (expressing).
The functional ability that these entities should have are:
- the region between Rec recombinase site sequence is inverted at each cell division.
- the protein A and protein B degradation rates should be fast enough to degrade them rapidly when not expressed.
- the protein A and protein B effects on (respectively) the promoter A and B expression capacity should be strong.
- promoter P should be strong in its constitutive expression.
The physical configuration:
The design relies on the alternate expression of the two strands of the DNA segment in between the Recombinase sites. Promoter P is placed outside the "flipping region" and controls its downstream region in the upper strand. The output of the device is given by the PoPS activity on the upper strand downstream from the second recombinase site. Since the region in between the recombinase sites flips, the output is controlled alternatively by the two DNA strands. In one strand the parts sequence is: repressor A, terminator, Promoter A. In the other the sequence is: repressor B, terminator, Promoter B.
The dynamics:
As cell division occurs, DNA enclosed by two inverted Recombinase sites is inverted. Initially the device is in the configuration with Repressor A and Promoter A on the upper strand. There is no Repressor A present in the cell, so Promoter A produces a constant PoPS output. As time passes, Repressor A accumulates and 'turns off' Promoter B. Meanwhile Repressor B is no produced and degrades so that, at the next division, Promoter B can produce PoPS output and the process repeats. This mechanism can be seen as the sequence of three phases: Forward, Flipping and Backward. We explain for each phases the dynamics:
Implementation
In the previous section we define which functional components are needed at the level of parts. In this section the members with biology competences evaluate the design and identify the actual biological parts that can be used. Further on, the DNA abstraction level details are proposed.
| Part abstraction level (Implementation) |
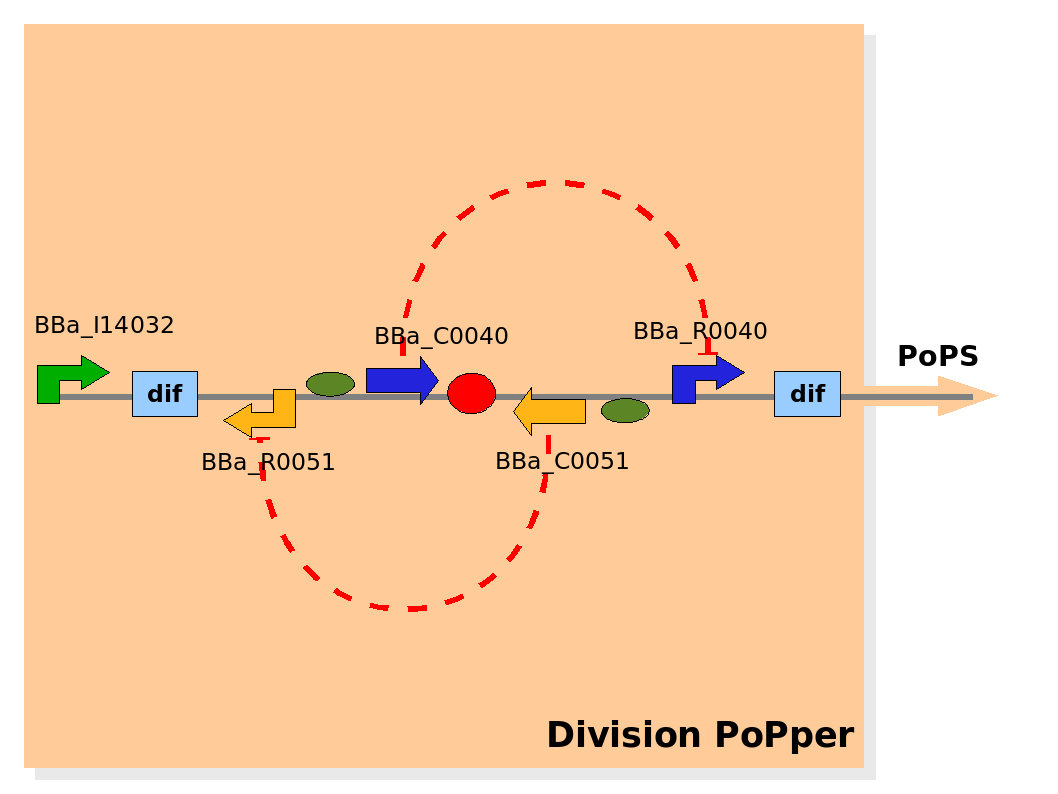 Implementation of the part structure in the Division PoPper device. Implementation of the part structure in the Division PoPper device.
|
Part abstraction level
The Division PoPper parts are:
- two Dif recombinase sites (design: Rec).
- Tet promoter (design: promoter A).
- TetR repressor (design: repressor A).
- CLambda promoter (design: promoter B).
- CLambda repressor (design: repressor B).
- Lac promoter (design: promoter P).
The Registry of Standard Parts contains already some of the elements we need, for the others we constructed and added their BioBricked version.
The Division PoPper parts registry entries are:
- two Dif recombinase sites: [http://partsregistry.org/Part:BBa_I742101 BBa_I742101],[http://partsregistry.org/Part:BBa_I742102 BBa_I742102]
- Tet promoter:[http://partsregistry.org/Part:BBa_R0040 BBa_R0040]
- TetR repressor:[http://partsregistry.org/Part:BBa_C0040 BBa_C0040]
- CLambda promoter:[http://partsregistry.org/Part:BBa_R0051 BBa_R0051]
- CLambda repressor:[http://partsregistry.org/Part:BBa_C0051 BBa_C0051]
- Lac promoter:[http://partsregistry.org/Part:BBa_I14032 BBa_I14032]
Click on the links for details.
The assumptions regarding the biological elements are:
- The Dif sites flip only once during each cell division
- TetR and ClambdaR are produced and degrade faster than the cell cycle
For this device to work, dif site-enclosed DNA needs to flip only once per division. Like all recombination sites, dif sites are directional and bacteria use directly repeated dif sites to resolve genome dimers. Research shows that this resolution occurs only at septation, and we use this temporal control to inverse a plasmid-borne sequence to induce different functions. For more information look at the Background section.
The Biology Processes: We used the Edinburgh Pathway Notation, a state transition graphical notation to express the biological processes of the Division PoPper. The details regarding the notations can be found in the paper [http://www-bm.ipk-gatersleben.de/stable/php/journal/articles/pdf/jib-36.pdf A Graphical Notation to Describe the Logical Interactions of Biological Pathways].
| DNA strands configuration |
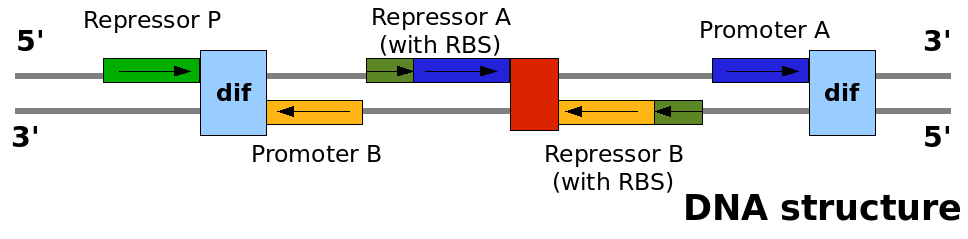 This graph explain on which DNA strand the parts are placed and with which coding direction. This graph explain on which DNA strand the parts are placed and with which coding direction.
|
DNA abstraction level
In order for this construct to be implemented as BioBricks combined in the normal fashion, it was necessary for some of the components to be synthesised in inverse orientation. The final list of components for the sequence, in order, is as follows:
- lac promoter [http://partsregistry.org/Part:BBa_I14032 BBa_I14032]
- dif site in forward orienation, generated by us as [http://partsregistry.org/Part:BBa_I742101 BBa_I742101]. (physically generated as a BioBrick and deposited).
- reverse version of lambda cI-controlled promoter [http://partsregistry.org/Part:BBa_R0051 BBa_R0051]: designed as [http://partsregistry.org/Part:BBa_I742126 BBa_I742126].
- ribosome binding site [http://partsregistry.org/Part:BBa_B0034 BBa_B0034].
- Tet repressor coding sequence with rapid degradation tag [http://partsregistry.org/Part:BBa_C0040 BBa_C0040].
- bidirectional terminator [http://partsregistry.org/Part:BBa_B0011 BBa_B0011] (initially it was planned to use a combination of several terminators here in different orientations, but after consultation with DNA synthesis company GeneArt, it was considered that the repeated sequences would cause difficulties in synthesis. If termination appears to be insufficient, sub-regions of the device can be extracted as separate biobricks by PCR, and further terminators inserted). In the final design submited to GeneArt, for certain reasons an inverse version of BBa_B0011 was used, and this was entered in our sandbox as [http://partsregistry.org/Part:BBa_I742129 BBa_I742129].
- Reverse coding sequence of lambda cI repressor BBa_C0051, entered in our sandbox as [http://partsregistry.org/Part:BBa_I742131 BBa_I742131].
- Reverse ribosome binding site BBa_B0034, entered in our sandbox as [http://partsregistry.org/Part:BBa_I742130 BBa_I742130]. Since the normal abbreviated biobrick scar between RBS and coding sequence could not be used in the inverse orientation, the length of the RBS was decreased by two bases to maintain the proper spacing.
- Tet repressor-controlled promoter [http://partsregistry.org/Part:BBa_R0040 BBa_R0040].
- dif site in reverse orientation, generated as [http://partsregistry.org/Part:BBa_I742102 BBa_I742102] and deposited.
- The total construct has been entered in our sandbox as [http://partsregistry.org/Part:BBa_I742132 BBa_I742132].
In order for the device to be tested, it will be necessary to insert it into a single copy BioBrick vector, since if multiple copies are present the likelihood is increased that not all will be in the same orientation, leading to simultaneous production of both repressors and lack of output (this cross-talk between separate instances of the device within a single system is a characteristic feature of biological systems which must always be considered) . For this purpose, the single copy F'-episome-based plasmid pHR277 (kanR) was generously provided by Professor Millicent Masters, Institute of Cell Biology, University of Edinburgh. This plasmid possesses unique XbaI and PstI sites which can be used to insert a biobrick; unfortunately, it also possesses an EcoRI site elsewhere on the plasmid so, unless this is removed, it cannot be converted into a fully compliant vector which can be used to assemble new BioBricks (in contrast to our Gram positive BioBrick vector pTG262-BB (BBa_I742123), described elsewhere in this wiki).
Proofs of Concept
The complex structure of the Division PoPper and the small amount of time we have for the construction of the device in the context of the iGEM competition led us to plan the construction of a simpler construct: the Proof of Concept device. The aim of the Proof of Concept device is to test the main assumptions at the basis of the Division PoPper device: that the recombinase mechanism is activated only once and at cell division and that the time between divisions is sufficient for the repression mechanism to be activated and inactivated. We planned to construct two experiments:
Experiment 1:
This experiment will prove whether or not the DNA between the two dif sites flips at all during cell division. If the a flip occurs, then the direction the promoter operates will be changed and YFP will be expressed. The BioBrick used for YFP expression is [http://partsregistry.org/Part:BBa_E0430 BBa_E0430]. The reverse lac promoter has been constructed and deposited as [http://partsregistry.org/Part:BBa_I742124 BBa_I742124].
Experiment 2:
This investigates the number of flips that occurs during division and will require rapidly degrading fluorescent proteins. After each division we expect to see a change in colour as the promoter activates a different reporter. We developed some mathematical models of this scenario that can be seen in the Modelling section. In the Wet Lab section you can find all the detail regarding the construction in the lab.