BerkiGEM2007Present1
From 2007.igem.org
JCAnderson (Talk | contribs) |
|||
| (67 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | == | + | <html> |
| + | <style> | ||
| + | #apDiv3 { | ||
| + | position:absolute; | ||
| + | left:10px; | ||
| + | top:-108px; | ||
| + | width:101px; | ||
| + | height:87px; | ||
| + | z-index:900; | ||
| + | } | ||
| + | </style> | ||
| + | <body> | ||
| + | <div id="apDiv3"><a href="https://2007.igem.org/Main_Page"><img name="Coverlogo" src="https://static.igem.org/mediawiki/2007/0/0e/Coverlogojpeg.jpg" width="101" height="87" alt=""></div></a> | ||
| + | </body> | ||
| + | </html> | ||
| + | <html> | ||
| + | <head> | ||
| + | <meta http-equiv="Content-Type" content="text/html; charset=utf-8" /> | ||
| + | <title>Untitled Document</title> | ||
| + | <style type="text/css"> | ||
| + | <!-- | ||
| + | #apDiv1 { | ||
| + | position:absolute; | ||
| + | left:2.5%; | ||
| + | top:50px; | ||
| + | width:95%; | ||
| + | height:90%; | ||
| + | z-index:1; | ||
| + | background-color: #FFFFFF; | ||
| + | } | ||
| + | body { | ||
| + | background-image: url(); | ||
| + | background-color: #FFFFFF; | ||
| + | background-repeat: repeat; | ||
| + | } | ||
| + | a:link { | ||
| + | color: #FF0000; | ||
| + | } | ||
| + | a:visited { | ||
| + | color: #990000; | ||
| + | } | ||
| + | a:hover { | ||
| + | color: #FF0000; | ||
| + | } | ||
| + | a:active { | ||
| + | color: #FF0000; | ||
| + | } | ||
| + | h1,h2,h3,h4,h5,h6 { | ||
| + | font-weight: bold; | ||
| + | } | ||
| + | h1 { | ||
| + | font-size: xx-large; | ||
| + | color: #000000; | ||
| + | } | ||
| + | h2 { | ||
| + | font-size: x-large; | ||
| + | color: #000000; | ||
| + | } | ||
| + | h3 { | ||
| + | font-size: large; | ||
| + | color: #000000; | ||
| + | } | ||
| + | --> | ||
| + | </style> | ||
| + | </head> | ||
| - | + | <body> | |
| - | ''' | + | <div align="center"><a href="https://2007.igem.org/Berkeley_UC"><<< Return to UC Berkeley iGEM 2007 | </a><a href="https://2007.igem.org/BerkiGEM2007Present4">Next Section: Chassis>></a></div> |
| - | + | <p align="center"> </p> | |
| - | + | <h1 align="center">Engineering Bactoblood for Oxygen Transport<br /> | |
| - | + | </h1> | |
| - | + | <p align="justify">The primary function of human erythrocytes is to transport oxygen to the body's tissues and remove CO2. This is accomplished principly by high concentrations of the protein hemoglobin. However, functional expression of hemoglobin requires the coexpression of the small molecule (heme) that specifically binds oxygen, proteins that promote the expression, folding, and addition of heme to hemoglobin, and proteins that maintain the oxidation state of hemoglobin and prevent the accumulation of toxic oxidizing species in the cell. Bactoblood will similarly require these activities, so we designed a hierarchical genetic device that encoded this oxygen transport function. Our design contains a heme biosynthesis devices, a hemoglobin generation device, a chaperone device, and a detoxifying device. Additionally, we investigated alternatives to hemoglobin that may provide superior oxygen transport to Bactoblood than their human counterpart.</p> | |
| - | + | <p align="justify"> </p> | |
| - | + | <h2 align="center">The Hemoglobin Generating Device</h2> | |
| - | The biosynthetic pathway | + | <p align="center"><img src="https://static.igem.org/mediawiki/2007/d/d1/Berk-Figure-HbA-HbB.png" name="HemoglobinCassette" width="337" height="111" id="HemoglobinCassette" /><img src="https://static.igem.org/mediawiki/2007/9/9e/Berk-Figure-Map.png" width="251" height="115" alt=""></p> |
| - | + | <h3 align="center">Overview: Human Hemoglobin A and Methionine Aminopeptidase</h3> | |
| - | + | <p align="justify">The primary component needed for efficient oxygen transport in our system is human hemoglobin A. (HbA) HbA is a tetramer that consists of two different subunits, α2β2. We constructed a device that expresses both the alpha (HbA) and beta (HbB) subunits of human hemoglobin A, under the control of a T7 promoter. We also created a similar device that will express mutant versions of human hemoglobin. To cleave the extra methionine residue that is present when expressed in prokaryotic cells, we have also constructed a device which expresses the Map gene, which encodes for methionine aminopeptidase (MetAP). By expressing both of these devices in our system, we achieve the expression of unmodified, fully functional adult human hemoglobin. </p> | |
| - | + | <h3 align="center">Oxygen Binding Affinity and P50</h3> | |
| - | + | <p align="justify">The first problem to be addressed is the insufficiently low P50 of wild type human hemoglobin. The P50 is the partial pressure of oxygen needed for 50% saturation. A low P50 means that the oxygen affinity is too high, which inhibits the ability of the hemoglobin to deliver oxygen to the needed tissues. The P50 for wild type human hemoglobin is ~3.8 torr under normal physiological conditions, however this varies with temperature and pH.</p> | |
| - | + | <p align="justify"><img src="https://static.igem.org/mediawiki/2007/e/e6/P50Comparison.gif" width="644" height="439" align="left"></p> | |
| - | + | <p align="justify">Demonstrated by George J. Brewer's image to the left, an increase in P50 is the same as shifting the oxygen binding curve to the right. A shift to the right is shown to increase the oxygen transport, thus a higher P50 is desirable for efficient oxygen transport. </p> | |
| - | + | <p align="justify"> </p> | |
| - | + | <p align="justify">In human erythrocytes, the oxygen binding affinity is decreased by the presence of an allosteric modifiers, primarily 2,3-diphosphoglycerate (2,3-DPG), which forces the hemoglobin conformation into the lower affinity deoxy state, or the T-state. By pushing the hemoglobin into the T-state, 2,3-DPG is effectively pushing any bound oxygen out from the heme center. This effectively lowers the oxygen binding affinity of the hemoglobin and increases the P50. An increase of 0.4mM in DPG concentration decreases oxygen affinity by about 1.0 torr. Since the normal concentration of DPG in erythrocytes is ~5mM, this raises the P50 to ~16.3 torr. </p> | |
| - | + | <p align="justify"> </p> | |
| - | + | <p align="justify">DPG isn't the only allosteric modifier of hemoglobin, to a lesser extent, various ions bind to a region near the N-terminal end of the hemoglobin protein and act to increase the P50 as well. In recombinant hemoglobin expressed in E. coli, there is an extra methionine residue at the N-terminal region of expressed proteins. For the case of hemoglobin, that extra methionine acts to inhibit the allosteric effects of certain ions. </p> | |
| - | + | <p align="justify"> </p> | |
| - | + | <h3 align="center">Wild Type and Mutant Hemoglobins<br /> | |
| - | + | </h3> | |
| - | + | <p align="justify">In our system, we have chosen to use well studied mutants of human hemoglobin which have been engineered to be permanently in the deoxy T-state, and the wild type human hemoglobin. The wild type is to compare the mutant hemoglobins to. The first generation Bactoblood blood substitute will use a hemoglobin mutant which has two mutations applied to the beta subunit. The mutations are named Presbyterian (beta-Asn108Lys) and providence (beta-Lys82Asp). It has been reported that human hemoglobin with these two point mutations has a P50 in the range of ~44 torr, an acceptable P50 value for a good blood substitute.</p> | |
| - | + | <p align="justify">The western blot to the bottom shows various mutant and wild type hemoglobin expressed under a Ptrc promoter. We constructed and tested different constructs to determine optimal hemoglobin yield. A goat-anti-human polyclonal antibody was used to identify the produced hemoglobin. All of these samples had heme added exogenously to the culture during growth and were allowed to grow overnight. The bands around 16kD are consistent with monomeric alpha and beta subunits of hemoglobin. Some of the constructs contain a fusion of alpha subunits with a glycine linker. More about this will be discussed in the hemoglobin solubility section. The western demonstrates that hemoglobin is being produced in our cells to a significant degree. In our potential final product, the heme will be produced in vivo and our cultures will be concentrated. This will greatly increase the concentration of hemoglobin in our cells to hopefully near physiological concentrations. </p> | |
| - | + | <h3 align="center"><img src="https://static.igem.org/mediawiki/2007/b/bc/Westernpicture.jpg" width="725" height="871" border="0" align="middle"></h3> | |
| - | + | <h3 align="center"><img name="" src="https://static.igem.org/mediawiki/2007/5/53/Lysis.gif" width="458" height="333" alt=""></h3> | |
| - | + | <h3 align="center"> </h3> | |
| - | + | <h3 align="center">Methionine Aminopeptidase</h3> | |
| - | + | <p align="justify">In prokaryotic cells, the N-terminal end of proteins retain the initial methionine residue from translation. In order to create human hemoglobin in the same form as it exists in erythrocytes, this extra methionine needs to be cleaved. The enzyme that does this occurs natrually in prokaryotes and is called methionine aminopeptidase (MetAP), encoded for by the Map gene. However, recombinant hemoglobin largely retains this extra methionine residue. By overexpressing metAP, it has been shown that a large percentage of the produced hemoglobin can be free of this extra methionine residue. It has also been shown that removal of the extra methionine residue increases the P50 slightly.</p> | |
| - | + | <p align="justify"> </p> | |
| - | + | <h2 align="center">The Chaperone Device</h2> | |
| - | + | <p align="center"><img src="https://static.igem.org/mediawiki/2007/a/aa/Berk-Figure-AHSP.png" width="257" height="122" /></p> | |
| - | + | <h3 align="center">Overview: Alpha Hemoglobin Stabilizing Protein</h3> | |
| - | + | <p>Alpha hemoglobin stabilizing protein (AHSP) is a chaperone protein normally found in erythrocytes. Because AHSP is important in the proper folding of hemoglobin, we created a device under a T7 promoter which will express AHSP in Bactoblood. </p> | |
| - | + | <h3 align="center"><img src="https://static.igem.org/mediawiki/2007/b/b0/AHSPreaction.png" width="636" height="476" align="left"></h3> | |
| - | + | <h3 align="center">Hemoglobin Solubility</h3> | |
| - | == | + | <p>The alpha subunit of hemoglobin is more prone to precipitation because an alpha-alpha dimer is insoluble under normal conditions. This can cause an excess of beta subunits and decreased output of functional tetrameric hemoglobin. In order to prevent alpha subunits from binding to themselves and precipitating out of solution, human erythrocytes contain an alpha hemoglobin stabilizing protein (AHSP), which acts as a chaperone. AHSP has the ability to bind to the alpha subunit of hemoglobin while keeping it soluble. The AHSP bound alpha subunit readily gives up its AHSP for a beta subunit which then goes on to form functional tetrameric hemoglobin. By expressing AHSP in Bactoblood, we expect to increase the yield of functional, soluble tetrameric hemoglobin. The mechanism for AHSP is shown in the figure to the left. </p> |
| - | + | <p><br />In addition, we have also explored a fusion of di-alpha subunits with a glycine linker. This has been shown to give a higher soluble output by stabilizing the alpha subunits and preventing precipitation. </p> | |
| - | + | <p align="justify"> </p> | |
| - | + | <p align="justify"><br /> | |
| - | + | </p> | |
| - | + | <h2 align="center">Heme Biosynthesis Device</h2> | |
| - | + | <p align="center"><img src="https://static.igem.org/mediawiki/2007/f/fd/Berk-Figure-hemABCD.png" width="563" height="122" /></p> | |
| - | The second gene | + | <h3 align="center">Overview: Heme</h3> |
| - | + | <p align="justify">Heme is a prosthetic group to hemoglobin. Heme consists of an iron atom surrounded by a porphyrin ring. Each hemoglobin tetramer is capable of binding up to four heme groups. One of the most important functions of heme, and the fuction we are interested in, is to assist in the transportation of diatomic gases, namely oxygen. The heme biosynthesis pathway is already present in E. coli, however, to achieve the high concentrations of functional hemoglobin needed for Bactoblood, we need lots of heme. We have constructed a device containing thefour genes hemA, hemB, hemC, and hemD. These genes are the primary bottlenecks in the heme biosynthesis pathway. We hope to greatly increase the production of heme by overexpressing these four genes under a T7 promoter. </p> | |
| - | + | <h3 align="center"><strong>Heme Biosynthesis Pathway</strong> </h3> | |
| - | + | <p>The biosynthetic pathway contains primarily eight enzymes. We included hemA (Delta-aminolevulinate synthase), hemB (Delta-aminolevulinic acid dehydratase), hemC (porphobilinogen deaminase), and hemD (uroporphyrinogen III synthase) in our system. These genes overproduce <em>precursors</em> to heme in our cells because over-accumulation of heme itself would result in toxicity. After successful subcloning experiments, the bacterial cell pellets would become reddish-brown due to the accumulation of porphyrins and heme.</p> | |
| - | + | <p><img src="https://static.igem.org/mediawiki/2007/0/01/96wellplateheme.jpg" width="498" height="308" align="left"></p> | |
| + | <h3 align="center">Construction of the Heme Device</h3> | ||
| + | <p>The hemA gene was cloned from both R.capsulatus and CFT073. Both were cloned to figure out which version would give a greater yield of heme precursors. We also cloned hemB, hemC, and hemD from MG1655. We attached single ribosomal binding sites to the genes, and we also attached a library of ribosomal binding sites to the genes. We used a library of ribosome binding sites so that we could grow up many clones in a 96 well plate and determine which ribosome binding sites were the strongest. The image of such a 96 well plate is shown above. Because color change is a phenotype of porphyrins and heme, it was easy to select single colonies that corresponded to the stronger ribosomal binding sites in library. These stronger clones would yield the greatest amount of heme and heme precursors and have the deepest red/brown color of all the clones. </p> | ||
| + | <p><br> | ||
| + | </p> | ||
| + | <p> </p> | ||
| + | <h3 align="center"><img src="https://static.igem.org/mediawiki/2007/9/9b/Hemespec.jpg" width="681" height="426" align="right"></h3> | ||
| + | <h3 align="center">Verification of the Heme Device</h3> | ||
| + | <p>With our construct complete, we employed UV-Vis to confirm that the red product we are seeing is, in fact, heme. The maximum absorbance for heme occurs at 412 nm. The graph above verifies that by alleviating the bottlenecks in the heme biosynthesis pathway, we have increased the concentration of heme in our system.</p> | ||
| + | <p> </p> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2007/0/0b/Bactoadcropped.gif" alt="" width="241" height="210" align="middle"></p> | ||
| + | <p> </p> | ||
| + | <p> </p> | ||
| + | <h2 align="center">Autoxidation Control<br /> | ||
| + | </h2> | ||
| + | <p align="center"><img src="https://static.igem.org/mediawiki/2007/7/7b/Berk-Figure-Cytochrome.png" width="582" height="112" /><img src="https://static.igem.org/mediawiki/2007/4/40/Berk-Figure-SodC-katG.png" width="294" height="105" /></p> | ||
| + | <h3 align="center">Overview: Cytochromes and Antioxidants</h3> | ||
| + | <p align="justify">During the process of binding and unbinding oxygen, the heme groups of the hemoglobin may spontaneously undergo autoxidation, ultimately causing the formation of damaging free radicals and changing the form of the iron in the heme center. To remedy these problems we created an antioxidant device and a methemoglobin reductase device. The antioxidant device contains the genes that express superoxide dismutase and catalase. The methemoglobin reductase device contains the genes that express cytochrome b5 and cytochrome b5 reductase. These two devices will keep our hemoglobin functional and our cells healthy. </p> | ||
| + | <h3 align="center">Autoxidation and Free Radical Production</h3> | ||
| + | <p align="justify">There are two problems to address in keeping our cells happy and our hemoglobin functional. The first problem is the spontaneous autoxidation of the heme centers in hemoglobin. During the binding and unbinding of oxygen, electrons are transfered back and forth between the iron center of heme and the bound oxygen molecule. Every once in a while, an electron will transfer to the oxygen and the oxygen will then unbind from the iron center and take the electron away from the iron. This creates a superoxide and a ferric Fe3+ heme. The superoxide eventually will degrate into a hydrogen peroxide, then into a hydroxyl radical, which is very toxic to the cell. The second problem is that the ferric heme can no longer bind oxygen, leaving the hemoglobin non-functional, this form is also known as methemoglobin. This autoxidation occurs on the order of hours, therefore accumulation of non-functional methemoglobin is significant if left alone. </p> | ||
| + | <h3 align="center"><img src="https://static.igem.org/mediawiki/2007/8/87/Antioxidantreactions.jpg" width="447" height="336" align="left"></h3> | ||
| + | <h3 align="center">Antioxidants</h3> | ||
| + | <p align="justify">Erythrocytes remedy the problem of free radical accumulation and damage by containing the antioxidant enzymes catalase and superoxide dismutase. These enzymes catalyze the breakdown of superoxide into oxygen and H2O. These enzymes are already used in E. Coli because free radicals are formed regularly during metabolism. However, we feel that because of our unusual accumulation of heme and hemoglobin, we must supplement the breakdown of the formed superoxides by overexpressing the catalase and superoxide dismutase genes in our system. The reaction mechanism to the left shows the pathway of superoxide degredation and the roles of superoxide dismutase and catalase. The desired breakdown of superoxide is through the two enzymes mentioned. The alternative pathway results in more non-functional ferric iron.</p> | ||
| + | <p align="justify"><img src="https://static.igem.org/mediawiki/2007/b/b4/Antioxidantdata.jpg" alt="" width="932" height="610" align="middle">The graph above shows data from our antioxidant assay. Paraquot, a substance that results in the formation of superoxide, was added to cells expressing the antioxidant enzymes. The data shows that the presence of both genes causes the cells to grow to a lower density than with only a single gene. This is unexpected and may be due to an unanticipated load brought onto the cells by overexpression of so much protein under T7 regulation. Further testing will be needed to confirm this. </p> | ||
| + | <h3 align="center"><img src="https://static.igem.org/mediawiki/2007/3/3c/Cytochromereactions.jpg" width="500" height="426" align="right"></h3> | ||
| + | <h3 align="center">Cytochrome b5 / Cytochrome b5 Reductase and Methemoglobin</h3> | ||
| + | <p align="justify"> Human erythrocytes have addressed the accumulation of methemoglobin caused by the autoxidation problem by using the NADH dependent enzyme, cytochrome b5 reductase. Working together with cytochrome b5, these two components allow for reduction of the methemoglobin heme centers back into its functional ferrous (Fe2+) form. The mechanism is shown to the right. The functional Fe3+ form of hemoglobin is said to have a dark chocolate color. </p> | ||
| + | <p align="justify"> </p> | ||
| + | <p align="center"><img src="https://static.igem.org/mediawiki/2007/7/7b/Cultures.jpg" width="518" height="289" alt=""></p> | ||
| + | <p align="center"><img src="https://static.igem.org/mediawiki/2007/c/cd/Pellets.jpg" width="517" height="290" alt=""></p> | ||
| + | <p align="justify">Shown above are cultures and pellets for the following constructs:<br> | ||
| + | Left: Mutant hemoglobin + hemA/hemB/hemC/hemD + cytochrome b5/cytochrome b5 reductase,<br> | ||
| + | Middle: hemA/hemB/hemC/hemD,<br> | ||
| + | Right: DH10B Negative control. <br> | ||
| + | It is significant to note that the color of the culture varies depending on multiple conditions which we have not yet fully identified.</p> | ||
| + | <p align="justify"> </p> | ||
| + | <h2 align="center">Hemoglobin Alternatives</h2> | ||
| + | <p align="justify">We also investigated two alternatives to the hemoglobin part in our device: H-NOX and Myoglobin. Although the intrinsic oxygen-carrying ability of these proteins is different from Hemoglobin, variants of these proteins have been engineered with similar P50 values. These variants might allow Bactoblood to carry more oxygen than hemoglobin. </p> | ||
| + | <p align="justify">H-NOX is a heme-based sensor that is found in bacteria. H-NOX is able to bind Oxygen using a distal pocket tyrosine. For this gene I added the T7 promoter we created for this project, an RBS site, and lastly a Bca1092 terminator. When we assayed this part the results were inconclusive. The part was assembled correctly but the assay didn't show strong signs of expression. </p> | ||
| + | <p align="justify">The second gene we explored was Sperm Whale Myoglobin. Myoglobin is a monomeric protein that behaves as an intracellular oxygen storage site. Sperm whale myoglobin in particular is easily found in large amounts in the whale's muscle tissue. The construction of this part was very similar to that of the H-NOX composite part. It used the same promoter, terminator, and RBS. The assay for Myoglobin showed a bit more promise but we couldn't conclusively show that Myoglobin was functionally expressed. </p> | ||
| + | <p align="center"> </p> | ||
| + | <div align="center"> | ||
| + | <h3>References</h3> | ||
| + | <p align="left">SJ Hoffman et al, “Expression of Fully Functional Tetrameric Human Hemoglobin in Escherichia coli”, PNAS, 87, p. 8521-8525, 1990. </p> | ||
| + | <p align="left">George J Brewer, “2,3-DPG and erythrocyte oxygen affinity”, Annual Reviews, 7092, 1974.</p> | ||
| + | <p align="left">Michael J. Weickert et al, “A Mutation That Improves Soluble Recombinant Hemoglobin Accumulation in Escherichia coli in Heme Excess”, Applied and Environmental Microbiology, p. 640-647, 1999. </p> | ||
| + | <p align="left">Michael J. Weichert et al, “High-Fidelity Translation of Recombinant Human Hemoglobin in Escherichia Coli”, Applied and Environmental Microbiology, p.1589-1593, 1998. </p> | ||
| + | <p align="left">Josef T. Prchal and Xylina T. Gregg, "Red Cell Enzymes", Hematology, 2005.</p> | ||
| + | <p align="left">Lloyd E et al, "Recombinant human erythrocyte cytochrome b5", Biochemistry, sep. 27; 33(38):11432-7, 1994.</p> | ||
| + | <p align="left">Robert M. Winslow, “Blood Substitutes”, Academic Press, 2005.</p> | ||
| + | <p align="left">T Shen <em>et al</em>, “Production of Unmodified Human Adult Hemoglobin in Escherichia coli”, PNAS, Vol. 90, pp. 8108-8112, September 1993.</p> | ||
| + | <p align="left">SJ Hoffman <em>et al</em>, “Expression of Fully Functional Tetrameric Human Hemoglobin in Escherichia coli”, PNAS, Vol. 87, pp. 8521-8525, November 1990. <br> | ||
| + | </p> | ||
| + | <p align="left">Seong Tae Jeong et al. "Recombinant Hemoglobin (alpha29Leucine -> Phenylalanine, alpha96Valine -> Tryptophan, beta108Asparagine -> Lysine) Exhibits Low Oxygen Affinity and High Cooperativity Combined with Resistance to Autoxidation", Biochemistry, 38, 13433-13442, 1999. </p> | ||
| + | <p align="left">Clara Fronticelli et al, "Allosteric Modulation by Tertiary Structure in Mammalian Hemoglobins", The Journal of Biological Chemistry, Vol 270, No. 51, pp. 30588-30592, 1995</p> | ||
| + | <p align="left">Ronald A. Hernan et al, "Human Hemoglobin Expressin in <em>Escherichia coli</em>: Importance of Optimal Codon Usage", Biochemistry, 31, 8619-8628, 1992</p> | ||
| + | <p align="left">Liang Feng et al, "Molecular Mechanism of AHSP-Mediated Stabilization of alpha-Hemoglobin", Cell, Vol 199, 629-640, 2004</p> | ||
| + | <p align="left">Seok Joon Kwon et al, "High-Level Production of Porphyrins in Metabolically Engineered <em>Escherichia coli</em>: Systematic Extension of a Pathway Assembled from Overexpressed Genes Involved in Heme Biosynthesis", Applied and Environmental Microbiology, p. 4875-4883, 2003</p> | ||
| + | <p align="left">Bernard B. Keele et al, "Superoxide Dismutase from <em>Escherichia coli</em> B", The Journal of Biological Chemsitry, Vol 245, No. 29, 1970</p> | ||
| + | <p align="left">KR Imlay and JA Imlay, "Cloning and analysis of sodC, encoding the copper-zine superoxide dismutase of Escherichia coli", Journal of Bacteriology, Vol. 178, No. 9, 2564-2571, 1996</p> | ||
| + | <p align="left">Barbara L. Triggs-Raine et al, "Nucleotide Sequence of katG, Encoding Catalase HPI of <em>Escherichia coli</em>", Journal of Bacteriology, p. 4415-4419, 1988. </p> | ||
| + | <p align="left">BM Babior, "Superoxide: a two-edged sword", Brazillian Journal of Medical and Biological Research, 30, 141-155, 1997</p> | ||
| + | <p align="left">Komei Shirabe et al, "Expression of human erythrocyto NADH-cytochrome b5 reductase as an alpha-thrombin-cleavable fused protein in <em>Escherichia coli</em>", Biochemica et Biophysica Acta, 1008, 189-192, 1989. </p> | ||
| + | <p align="left">Debbie D.W Hwang et al, "Co-expression of gluthathionine S-transerase with methionine aminopeptidase: a system of producing enriched N-terminal processed proteins in <em>Escherichia Coli</em>", Biochem. J, 338, 335-342, 1999. </p> | ||
| + | <p align="left">Tong-Jian Shen et al, "Production of human normal adult and fetal hemoglobins in <em>Escherichia coli</em>", Protein Engineering, vol. 10, no. 9, pp 1085-1097, 1997. </p> | ||
| + | <p> </p> | ||
| + | <p><a href="https://2007.igem.org/Berkeley_UC"><<< Return to UC Berkeley iGEM 2007 | </a><a href="https://2007.igem.org/BerkiGEM2007Present4">Next Section: Chassis>></a></p> | ||
| + | </div> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 03:31, 27 October 2007
Engineering Bactoblood for Oxygen Transport
The primary function of human erythrocytes is to transport oxygen to the body's tissues and remove CO2. This is accomplished principly by high concentrations of the protein hemoglobin. However, functional expression of hemoglobin requires the coexpression of the small molecule (heme) that specifically binds oxygen, proteins that promote the expression, folding, and addition of heme to hemoglobin, and proteins that maintain the oxidation state of hemoglobin and prevent the accumulation of toxic oxidizing species in the cell. Bactoblood will similarly require these activities, so we designed a hierarchical genetic device that encoded this oxygen transport function. Our design contains a heme biosynthesis devices, a hemoglobin generation device, a chaperone device, and a detoxifying device. Additionally, we investigated alternatives to hemoglobin that may provide superior oxygen transport to Bactoblood than their human counterpart.
The Hemoglobin Generating Device


Overview: Human Hemoglobin A and Methionine Aminopeptidase
The primary component needed for efficient oxygen transport in our system is human hemoglobin A. (HbA) HbA is a tetramer that consists of two different subunits, α2β2. We constructed a device that expresses both the alpha (HbA) and beta (HbB) subunits of human hemoglobin A, under the control of a T7 promoter. We also created a similar device that will express mutant versions of human hemoglobin. To cleave the extra methionine residue that is present when expressed in prokaryotic cells, we have also constructed a device which expresses the Map gene, which encodes for methionine aminopeptidase (MetAP). By expressing both of these devices in our system, we achieve the expression of unmodified, fully functional adult human hemoglobin.
Oxygen Binding Affinity and P50
The first problem to be addressed is the insufficiently low P50 of wild type human hemoglobin. The P50 is the partial pressure of oxygen needed for 50% saturation. A low P50 means that the oxygen affinity is too high, which inhibits the ability of the hemoglobin to deliver oxygen to the needed tissues. The P50 for wild type human hemoglobin is ~3.8 torr under normal physiological conditions, however this varies with temperature and pH.

Demonstrated by George J. Brewer's image to the left, an increase in P50 is the same as shifting the oxygen binding curve to the right. A shift to the right is shown to increase the oxygen transport, thus a higher P50 is desirable for efficient oxygen transport.
In human erythrocytes, the oxygen binding affinity is decreased by the presence of an allosteric modifiers, primarily 2,3-diphosphoglycerate (2,3-DPG), which forces the hemoglobin conformation into the lower affinity deoxy state, or the T-state. By pushing the hemoglobin into the T-state, 2,3-DPG is effectively pushing any bound oxygen out from the heme center. This effectively lowers the oxygen binding affinity of the hemoglobin and increases the P50. An increase of 0.4mM in DPG concentration decreases oxygen affinity by about 1.0 torr. Since the normal concentration of DPG in erythrocytes is ~5mM, this raises the P50 to ~16.3 torr.
DPG isn't the only allosteric modifier of hemoglobin, to a lesser extent, various ions bind to a region near the N-terminal end of the hemoglobin protein and act to increase the P50 as well. In recombinant hemoglobin expressed in E. coli, there is an extra methionine residue at the N-terminal region of expressed proteins. For the case of hemoglobin, that extra methionine acts to inhibit the allosteric effects of certain ions.
Wild Type and Mutant Hemoglobins
In our system, we have chosen to use well studied mutants of human hemoglobin which have been engineered to be permanently in the deoxy T-state, and the wild type human hemoglobin. The wild type is to compare the mutant hemoglobins to. The first generation Bactoblood blood substitute will use a hemoglobin mutant which has two mutations applied to the beta subunit. The mutations are named Presbyterian (beta-Asn108Lys) and providence (beta-Lys82Asp). It has been reported that human hemoglobin with these two point mutations has a P50 in the range of ~44 torr, an acceptable P50 value for a good blood substitute.
The western blot to the bottom shows various mutant and wild type hemoglobin expressed under a Ptrc promoter. We constructed and tested different constructs to determine optimal hemoglobin yield. A goat-anti-human polyclonal antibody was used to identify the produced hemoglobin. All of these samples had heme added exogenously to the culture during growth and were allowed to grow overnight. The bands around 16kD are consistent with monomeric alpha and beta subunits of hemoglobin. Some of the constructs contain a fusion of alpha subunits with a glycine linker. More about this will be discussed in the hemoglobin solubility section. The western demonstrates that hemoglobin is being produced in our cells to a significant degree. In our potential final product, the heme will be produced in vivo and our cultures will be concentrated. This will greatly increase the concentration of hemoglobin in our cells to hopefully near physiological concentrations.
Methionine Aminopeptidase
In prokaryotic cells, the N-terminal end of proteins retain the initial methionine residue from translation. In order to create human hemoglobin in the same form as it exists in erythrocytes, this extra methionine needs to be cleaved. The enzyme that does this occurs natrually in prokaryotes and is called methionine aminopeptidase (MetAP), encoded for by the Map gene. However, recombinant hemoglobin largely retains this extra methionine residue. By overexpressing metAP, it has been shown that a large percentage of the produced hemoglobin can be free of this extra methionine residue. It has also been shown that removal of the extra methionine residue increases the P50 slightly.
The Chaperone Device
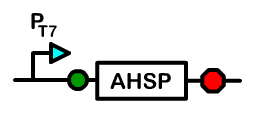
Overview: Alpha Hemoglobin Stabilizing Protein
Alpha hemoglobin stabilizing protein (AHSP) is a chaperone protein normally found in erythrocytes. Because AHSP is important in the proper folding of hemoglobin, we created a device under a T7 promoter which will express AHSP in Bactoblood.
Hemoglobin Solubility
The alpha subunit of hemoglobin is more prone to precipitation because an alpha-alpha dimer is insoluble under normal conditions. This can cause an excess of beta subunits and decreased output of functional tetrameric hemoglobin. In order to prevent alpha subunits from binding to themselves and precipitating out of solution, human erythrocytes contain an alpha hemoglobin stabilizing protein (AHSP), which acts as a chaperone. AHSP has the ability to bind to the alpha subunit of hemoglobin while keeping it soluble. The AHSP bound alpha subunit readily gives up its AHSP for a beta subunit which then goes on to form functional tetrameric hemoglobin. By expressing AHSP in Bactoblood, we expect to increase the yield of functional, soluble tetrameric hemoglobin. The mechanism for AHSP is shown in the figure to the left.
In addition, we have also explored a fusion of di-alpha subunits with a glycine linker. This has been shown to give a higher soluble output by stabilizing the alpha subunits and preventing precipitation.
Heme Biosynthesis Device

Overview: Heme
Heme is a prosthetic group to hemoglobin. Heme consists of an iron atom surrounded by a porphyrin ring. Each hemoglobin tetramer is capable of binding up to four heme groups. One of the most important functions of heme, and the fuction we are interested in, is to assist in the transportation of diatomic gases, namely oxygen. The heme biosynthesis pathway is already present in E. coli, however, to achieve the high concentrations of functional hemoglobin needed for Bactoblood, we need lots of heme. We have constructed a device containing thefour genes hemA, hemB, hemC, and hemD. These genes are the primary bottlenecks in the heme biosynthesis pathway. We hope to greatly increase the production of heme by overexpressing these four genes under a T7 promoter.
Heme Biosynthesis Pathway
The biosynthetic pathway contains primarily eight enzymes. We included hemA (Delta-aminolevulinate synthase), hemB (Delta-aminolevulinic acid dehydratase), hemC (porphobilinogen deaminase), and hemD (uroporphyrinogen III synthase) in our system. These genes overproduce precursors to heme in our cells because over-accumulation of heme itself would result in toxicity. After successful subcloning experiments, the bacterial cell pellets would become reddish-brown due to the accumulation of porphyrins and heme.

Construction of the Heme Device
The hemA gene was cloned from both R.capsulatus and CFT073. Both were cloned to figure out which version would give a greater yield of heme precursors. We also cloned hemB, hemC, and hemD from MG1655. We attached single ribosomal binding sites to the genes, and we also attached a library of ribosomal binding sites to the genes. We used a library of ribosome binding sites so that we could grow up many clones in a 96 well plate and determine which ribosome binding sites were the strongest. The image of such a 96 well plate is shown above. Because color change is a phenotype of porphyrins and heme, it was easy to select single colonies that corresponded to the stronger ribosomal binding sites in library. These stronger clones would yield the greatest amount of heme and heme precursors and have the deepest red/brown color of all the clones.
Verification of the Heme Device
With our construct complete, we employed UV-Vis to confirm that the red product we are seeing is, in fact, heme. The maximum absorbance for heme occurs at 412 nm. The graph above verifies that by alleviating the bottlenecks in the heme biosynthesis pathway, we have increased the concentration of heme in our system.

Autoxidation Control


Overview: Cytochromes and Antioxidants
During the process of binding and unbinding oxygen, the heme groups of the hemoglobin may spontaneously undergo autoxidation, ultimately causing the formation of damaging free radicals and changing the form of the iron in the heme center. To remedy these problems we created an antioxidant device and a methemoglobin reductase device. The antioxidant device contains the genes that express superoxide dismutase and catalase. The methemoglobin reductase device contains the genes that express cytochrome b5 and cytochrome b5 reductase. These two devices will keep our hemoglobin functional and our cells healthy.
Autoxidation and Free Radical Production
There are two problems to address in keeping our cells happy and our hemoglobin functional. The first problem is the spontaneous autoxidation of the heme centers in hemoglobin. During the binding and unbinding of oxygen, electrons are transfered back and forth between the iron center of heme and the bound oxygen molecule. Every once in a while, an electron will transfer to the oxygen and the oxygen will then unbind from the iron center and take the electron away from the iron. This creates a superoxide and a ferric Fe3+ heme. The superoxide eventually will degrate into a hydrogen peroxide, then into a hydroxyl radical, which is very toxic to the cell. The second problem is that the ferric heme can no longer bind oxygen, leaving the hemoglobin non-functional, this form is also known as methemoglobin. This autoxidation occurs on the order of hours, therefore accumulation of non-functional methemoglobin is significant if left alone.
Antioxidants
Erythrocytes remedy the problem of free radical accumulation and damage by containing the antioxidant enzymes catalase and superoxide dismutase. These enzymes catalyze the breakdown of superoxide into oxygen and H2O. These enzymes are already used in E. Coli because free radicals are formed regularly during metabolism. However, we feel that because of our unusual accumulation of heme and hemoglobin, we must supplement the breakdown of the formed superoxides by overexpressing the catalase and superoxide dismutase genes in our system. The reaction mechanism to the left shows the pathway of superoxide degredation and the roles of superoxide dismutase and catalase. The desired breakdown of superoxide is through the two enzymes mentioned. The alternative pathway results in more non-functional ferric iron.
 The graph above shows data from our antioxidant assay. Paraquot, a substance that results in the formation of superoxide, was added to cells expressing the antioxidant enzymes. The data shows that the presence of both genes causes the cells to grow to a lower density than with only a single gene. This is unexpected and may be due to an unanticipated load brought onto the cells by overexpression of so much protein under T7 regulation. Further testing will be needed to confirm this.
The graph above shows data from our antioxidant assay. Paraquot, a substance that results in the formation of superoxide, was added to cells expressing the antioxidant enzymes. The data shows that the presence of both genes causes the cells to grow to a lower density than with only a single gene. This is unexpected and may be due to an unanticipated load brought onto the cells by overexpression of so much protein under T7 regulation. Further testing will be needed to confirm this.
Cytochrome b5 / Cytochrome b5 Reductase and Methemoglobin
Human erythrocytes have addressed the accumulation of methemoglobin caused by the autoxidation problem by using the NADH dependent enzyme, cytochrome b5 reductase. Working together with cytochrome b5, these two components allow for reduction of the methemoglobin heme centers back into its functional ferrous (Fe2+) form. The mechanism is shown to the right. The functional Fe3+ form of hemoglobin is said to have a dark chocolate color.


Shown above are cultures and pellets for the following constructs:
Left: Mutant hemoglobin + hemA/hemB/hemC/hemD + cytochrome b5/cytochrome b5 reductase,
Middle: hemA/hemB/hemC/hemD,
Right: DH10B Negative control.
It is significant to note that the color of the culture varies depending on multiple conditions which we have not yet fully identified.
Hemoglobin Alternatives
We also investigated two alternatives to the hemoglobin part in our device: H-NOX and Myoglobin. Although the intrinsic oxygen-carrying ability of these proteins is different from Hemoglobin, variants of these proteins have been engineered with similar P50 values. These variants might allow Bactoblood to carry more oxygen than hemoglobin.
H-NOX is a heme-based sensor that is found in bacteria. H-NOX is able to bind Oxygen using a distal pocket tyrosine. For this gene I added the T7 promoter we created for this project, an RBS site, and lastly a Bca1092 terminator. When we assayed this part the results were inconclusive. The part was assembled correctly but the assay didn't show strong signs of expression.
The second gene we explored was Sperm Whale Myoglobin. Myoglobin is a monomeric protein that behaves as an intracellular oxygen storage site. Sperm whale myoglobin in particular is easily found in large amounts in the whale's muscle tissue. The construction of this part was very similar to that of the H-NOX composite part. It used the same promoter, terminator, and RBS. The assay for Myoglobin showed a bit more promise but we couldn't conclusively show that Myoglobin was functionally expressed.
References
SJ Hoffman et al, “Expression of Fully Functional Tetrameric Human Hemoglobin in Escherichia coli”, PNAS, 87, p. 8521-8525, 1990.
George J Brewer, “2,3-DPG and erythrocyte oxygen affinity”, Annual Reviews, 7092, 1974.
Michael J. Weickert et al, “A Mutation That Improves Soluble Recombinant Hemoglobin Accumulation in Escherichia coli in Heme Excess”, Applied and Environmental Microbiology, p. 640-647, 1999.
Michael J. Weichert et al, “High-Fidelity Translation of Recombinant Human Hemoglobin in Escherichia Coli”, Applied and Environmental Microbiology, p.1589-1593, 1998.
Josef T. Prchal and Xylina T. Gregg, "Red Cell Enzymes", Hematology, 2005.
Lloyd E et al, "Recombinant human erythrocyte cytochrome b5", Biochemistry, sep. 27; 33(38):11432-7, 1994.
Robert M. Winslow, “Blood Substitutes”, Academic Press, 2005.
T Shen et al, “Production of Unmodified Human Adult Hemoglobin in Escherichia coli”, PNAS, Vol. 90, pp. 8108-8112, September 1993.
SJ Hoffman et al, “Expression of Fully Functional Tetrameric Human Hemoglobin in Escherichia coli”, PNAS, Vol. 87, pp. 8521-8525, November 1990.
Seong Tae Jeong et al. "Recombinant Hemoglobin (alpha29Leucine -> Phenylalanine, alpha96Valine -> Tryptophan, beta108Asparagine -> Lysine) Exhibits Low Oxygen Affinity and High Cooperativity Combined with Resistance to Autoxidation", Biochemistry, 38, 13433-13442, 1999.
Clara Fronticelli et al, "Allosteric Modulation by Tertiary Structure in Mammalian Hemoglobins", The Journal of Biological Chemistry, Vol 270, No. 51, pp. 30588-30592, 1995
Ronald A. Hernan et al, "Human Hemoglobin Expressin in Escherichia coli: Importance of Optimal Codon Usage", Biochemistry, 31, 8619-8628, 1992
Liang Feng et al, "Molecular Mechanism of AHSP-Mediated Stabilization of alpha-Hemoglobin", Cell, Vol 199, 629-640, 2004
Seok Joon Kwon et al, "High-Level Production of Porphyrins in Metabolically Engineered Escherichia coli: Systematic Extension of a Pathway Assembled from Overexpressed Genes Involved in Heme Biosynthesis", Applied and Environmental Microbiology, p. 4875-4883, 2003
Bernard B. Keele et al, "Superoxide Dismutase from Escherichia coli B", The Journal of Biological Chemsitry, Vol 245, No. 29, 1970
KR Imlay and JA Imlay, "Cloning and analysis of sodC, encoding the copper-zine superoxide dismutase of Escherichia coli", Journal of Bacteriology, Vol. 178, No. 9, 2564-2571, 1996
Barbara L. Triggs-Raine et al, "Nucleotide Sequence of katG, Encoding Catalase HPI of Escherichia coli", Journal of Bacteriology, p. 4415-4419, 1988.
BM Babior, "Superoxide: a two-edged sword", Brazillian Journal of Medical and Biological Research, 30, 141-155, 1997
Komei Shirabe et al, "Expression of human erythrocyto NADH-cytochrome b5 reductase as an alpha-thrombin-cleavable fused protein in Escherichia coli", Biochemica et Biophysica Acta, 1008, 189-192, 1989.
Debbie D.W Hwang et al, "Co-expression of gluthathionine S-transerase with methionine aminopeptidase: a system of producing enriched N-terminal processed proteins in Escherichia Coli", Biochem. J, 338, 335-342, 1999.
Tong-Jian Shen et al, "Production of human normal adult and fetal hemoglobins in Escherichia coli", Protein Engineering, vol. 10, no. 9, pp 1085-1097, 1997.
<<< Return to UC Berkeley iGEM 2007 | Next Section: Chassis>>






