Tristable/Intro to Tristable
From 2007.igem.org
< Tristable(Difference between revisions)
| (5 intermediate revisions not shown) | |||
| Line 78: | Line 78: | ||
<tr> | <tr> | ||
<td width=99> | <td width=99> | ||
| - | <a href="Brown"><image src="https://static.igem.org/mediawiki/2007/2/20/Home_reg.gif" alt="Home" border=0></a> | + | <a href="../Brown"><image src="https://static.igem.org/mediawiki/2007/2/20/Home_reg.gif" alt="Home" border=0></a> |
</td> | </td> | ||
<td width=99> | <td width=99> | ||
| - | <a href="lead"><image src="https://static.igem.org/mediawiki/2007/2/2a/Lead_reg.gif" alt="Lead Sensor" border=0></a> | + | <a href="../lead"><image src="https://static.igem.org/mediawiki/2007/2/2a/Lead_reg.gif" alt="Lead Sensor" border=0></a> |
</td> | </td> | ||
<td width=99> | <td width=99> | ||
| - | <a href="tristable"><image src="https://static.igem.org/mediawiki/2007/7/79/Tri_pressed.gif" alt="Tristable Switch" border=0></a> | + | <a href="../tristable"><image src="https://static.igem.org/mediawiki/2007/7/79/Tri_pressed.gif" alt="Tristable Switch" border=0></a> |
</td> | </td> | ||
<td width=99> | <td width=99> | ||
| - | <a href="community"><image src="https://static.igem.org/mediawiki/2007/e/e7/Comm_reg.gif" alt="Community" border=0></a> | + | <a href="../community"><image src="https://static.igem.org/mediawiki/2007/e/e7/Comm_reg.gif" alt="Community" border=0></a> |
</td> | </td> | ||
<td width=99> | <td width=99> | ||
| - | <a href="supplemental"><image src="https://static.igem.org/mediawiki/2007/8/81/Supp_reg.gif" alt="Supplemental" border=0></a> | + | <a href="../supplemental"><image src="https://static.igem.org/mediawiki/2007/8/81/Supp_reg.gif" alt="Supplemental" border=0></a> |
</td> | </td> | ||
<td width=99> | <td width=99> | ||
| - | <a href="about"><image src="https://static.igem.org/mediawiki/2007/c/ca/About_reg.gif" alt="About us" border=0></a> | + | <a href="../about"><image src="https://static.igem.org/mediawiki/2007/c/ca/About_reg.gif" alt="About us" border=0></a> |
</td> | </td> | ||
<td></td> | <td></td> | ||
| Line 118: | Line 118: | ||
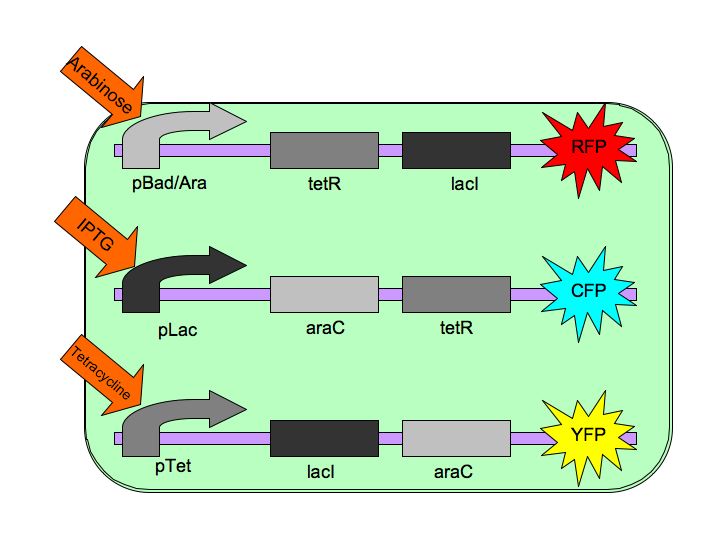
| - | Each of the three repressors are inactivated by one of three chemicals, the three inducer chemicals mentioned earlier. These three([http://en.wikipedia.org/wiki/Arabinose arabinose], [http://en.wikipedia.org/wiki/IPTG IPTG] (Isopropyl β-D-1-thiogalactopyranoside) and [http://en.wikipedia.org/wiki/Tetracycline Tetracycline], respectively), cause conformational changes in their respective repressor proteins which keeps them from binding to DNA in an inhibitory manner which leads to gene expression. For example, in the presence of arabinose, AraC cannot repress pAraC/BAD so LacI and TetR are produced which in turn repress pTet and pLac and the pAraC/BAD construct is turned on. | + | Each of the three repressors are inactivated by one of three chemicals, the three inducer chemicals mentioned earlier. These three([http://en.wikipedia.org/wiki/Arabinose L-arabinose], [http://en.wikipedia.org/wiki/IPTG IPTG] (Isopropyl β-D-1-thiogalactopyranoside) and [http://en.wikipedia.org/wiki/Tetracycline Tetracycline], respectively), cause conformational changes in their respective repressor proteins which keeps them from binding to DNA in an inhibitory manner which leads to gene expression. For example, in the presence of arabinose, AraC cannot repress pAraC/BAD so LacI and TetR are produced which in turn repress pTet and pLac and the pAraC/BAD construct is turned on. |
| Line 137: | Line 137: | ||
==LacI== | ==LacI== | ||
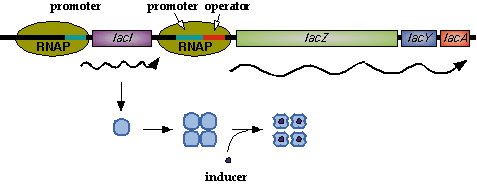
| - | In nature, LacI represses pLac which promotes the LacYZA genes that metabolize lactose. Thus LacI represses pLac except in the presence of lactose (or lactose mimics, eg IPTG). [[Image:LacI_repressor.gif|thumb|left|Image[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html]. LacI forms a tetramer and represses pLac. | + | In nature, LacI represses pLac which promotes the LacYZA genes that metabolize lactose. Thus LacI represses pLac except in the presence of lactose (or lactose mimics, eg IPTG). [[Image:LacI_repressor.gif|thumb|left|Image[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html]. LacI forms a tetramer and represses pLac. IPTG causes a conformation change in LacI.]] Lactose causes a conformational change which inhibits LacI from binding to the operator site of pLac. Four LacI proteins form a tetramer to inhibit pLac and four inducer molecules are required to cause the full conformational change in the repressor.[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html] |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| Line 150: | Line 155: | ||
==TetR== | ==TetR== | ||
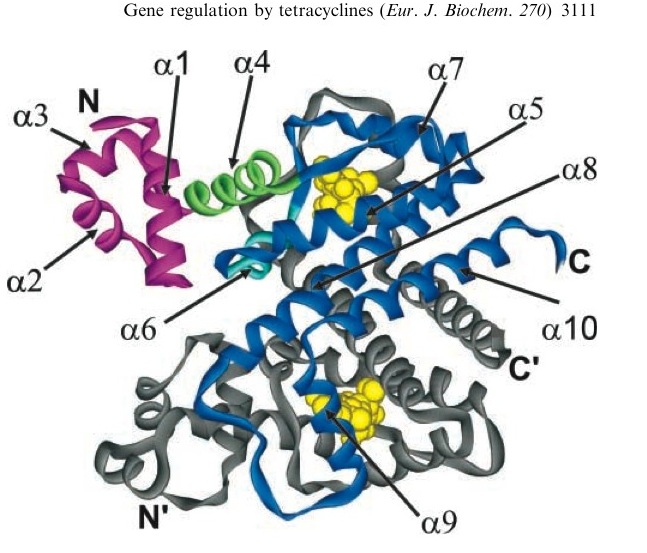
| - | TetR represses the constitutive promoter pTet. In the presence of tetracycline, an antibiotic, a conformational change in TetR inhibits the protein from binding to the operator region. In nature, pTet promotes TetR and TetA. The latter of which acts to pump tetracycline out of the cell, thus the pump is only activated in the presence of Tetracycline.[http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation] | + | TetR represses the constitutive promoter pTet. In the presence of tetracycline, an antibiotic, a conformational change in TetR inhibits the protein from binding to the operator region. In nature, pTet promotes TetR and TetA. The latter of which acts to pump tetracycline out of the cell, thus the pump is only activated in the presence of Tetracycline.[[Image:Tc_bound_to_TetR.jpg|thumb|left|A tetracycline molecule binds to each of the two TetR monomers to form a dimer]][http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation] |
The TetR, as it turns out is a very tight repressor and a range of 0 to 1 ug/ml has been shown to cause a 5 order of magnitude change in luciferase production.[http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1319065&query_hl=1&itool=pubmed_docsum] | The TetR, as it turns out is a very tight repressor and a range of 0 to 1 ug/ml has been shown to cause a 5 order of magnitude change in luciferase production.[http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1319065&query_hl=1&itool=pubmed_docsum] | ||
| - | + | ||
| + | |||
| + | We will be using anhydrotetracycline (aTc) as the inducer molecule as it has been shown to exhibit stronger binding to TetR and is not an antibiotic that will compromise our system. | ||
Latest revision as of 05:58, 25 October 2007