Security device
From 2007.igem.org
Eismoustique (Talk | contribs) |
Eismoustique (Talk | contribs) |
||
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | Back to [[Paris/Applications]] | + | Back to [[Paris/Applications|Perspectives]] |
| Line 8: | Line 8: | ||
* EVENT 1 | * EVENT 1 | ||
| - | In order to achieve massive induction of recombination, | + | In order to achieve massive induction of recombination, a dormant copy of Cre site specific recombinase could be added to the SMB system. This copy would be plasmid born & would placed under control of an inducible promoter. |
| - | But a certain proportion of G cells would | + | But a certain proportion of G cells would most certainly persist undifferentiated. |
* EVENT 2 | * EVENT 2 | ||
| - | The solution to second challenge could be coupling the high recombination switch to a | + | The solution to second challenge could be coupling the high recombination switch to a delayed suicide switch in the gem line cells. include the expression of ccdB suicide gene in the germ line cells. |
| - | + | ||
| - | + | ||
| + | This could be the opportunity to practically apply one of the founding principles of synthetic biology: modularity in systems assembly & organisation. | ||
Indeed, a time delay circuit has already been described, constructed & theoretically studied (Sara Hooshangi et al. 2005, Sara Hooshangi et al. 2006 and Juan M. Pedraza et al. 2006). | Indeed, a time delay circuit has already been described, constructed & theoretically studied (Sara Hooshangi et al. 2005, Sara Hooshangi et al. 2006 and Juan M. Pedraza et al. 2006). | ||
Using such a time delay circuit could allow using a single signal to induce event 1, followed after a delay by event 2. | Using such a time delay circuit could allow using a single signal to induce event 1, followed after a delay by event 2. | ||
| + | |||
| + | |||
In addition | In addition | ||
| - | Size based screening could be | + | Size based screening could be performed before environmental release. Indeed, S cells having lost ftsK expression present an interesting phenotype: they grow in size without division taking place, they "filament". The size threshold used in the cell sorting process should be determined by studying size distribution within germ cell and somatic cell populations. This could to improve security. <br> |
| Line 27: | Line 28: | ||
| - | + | In the figure above, the recombination procress accompanying differentiation would lead to the transcription of a ccdB lock in somatic cells. This should protect the somatic cells from the suicide event that germ line cells go through. This lock could be a post transcriptional regulatory mechanism as this class of regulation requires less latency time before appearing. | |
==References== | ==References== | ||
| + | |||
| + | ===Time delay circuit=== | ||
*'''Ultrasensitivity and noise propagation in a synthetic transcriptional cascade''' | *'''Ultrasensitivity and noise propagation in a synthetic transcriptional cascade''' | ||
| Line 43: | Line 46: | ||
Sara Hooshangi and Ron Weiss | Sara Hooshangi and Ron Weiss | ||
CHAOS 16, 026108 (2006) | CHAOS 16, 026108 (2006) | ||
| + | |||
| + | |||
| + | ===RNA based regulation in Synthetic Biology=== | ||
| + | |||
| + | *'''RNA synthetic biology'''. | ||
| + | Farren J Isaacs1, Daniel J Dwyer2,3 & James J Collins3. | ||
| + | NATURE BIOTECHNOLOGY Review VOLUME 24 NUMBER 5 MAY 2006. | ||
| + | |||
| + | |||
| + | '''Engineered riboregulators''' | ||
| + | |||
| + | *'''Engineered riboregulators enable post-transcriptional control of'''. | ||
| + | Isaacs, F.J. et al. | ||
| + | gene expression. Nat. Biotechnol. 22, 841–847 (2004). | ||
| + | |||
| + | |||
| + | '''Ribosome mRNA pairing''' | ||
| + | |||
| + | *'''network of orthogonal ribosome-mRNA pairs'''. | ||
| + | Rackham, O. & Chin, J.W. A. | ||
| + | Nat.Chem. Biol. 1, 159–166 (2005). | ||
| + | |||
| + | *'''Cellular logic with orthogonal ribosomes'''. | ||
| + | Rackham, O. & Chin, J.W. | ||
| + | J. Am. Chem. Soc. 127, 17584–17585 (2005). | ||
| + | |||
| + | |||
| + | '''Engineered ligand-controlled riboregulators (Riboswitches)''' | ||
| + | |||
| + | *'''Controlling gene expression in living cells through small | ||
| + | molecule-RNA interactions'''. | ||
| + | Werstuck, G. & Green, M.R. | ||
| + | Science 282, 296–298 (1998). | ||
| + | |||
| + | *'''theophylline responsive riboswitch based on helix slipping controls | ||
| + | gene expression in vivo.'''. | ||
| + | Suess, B., Fink, B., Berens, C., Stentz, R. & Hillen, W. A. | ||
| + | Nucleic Acids Res. 32, 1610–1614 (2004). | ||
| + | |||
| + | *'''Programmable ligand-controlled riboregulators of eukaryotic | ||
| + | gene expression'''. | ||
| + | Bayer, T.S. & Smolke, C.D. | ||
| + | Nat. Biotechnol. 23, 337–343 (2005). | ||
| + | |||
| + | Back to [[Paris/Applications|Perspectives]] | ||
Latest revision as of 23:59, 26 October 2007
Back to Perspectives
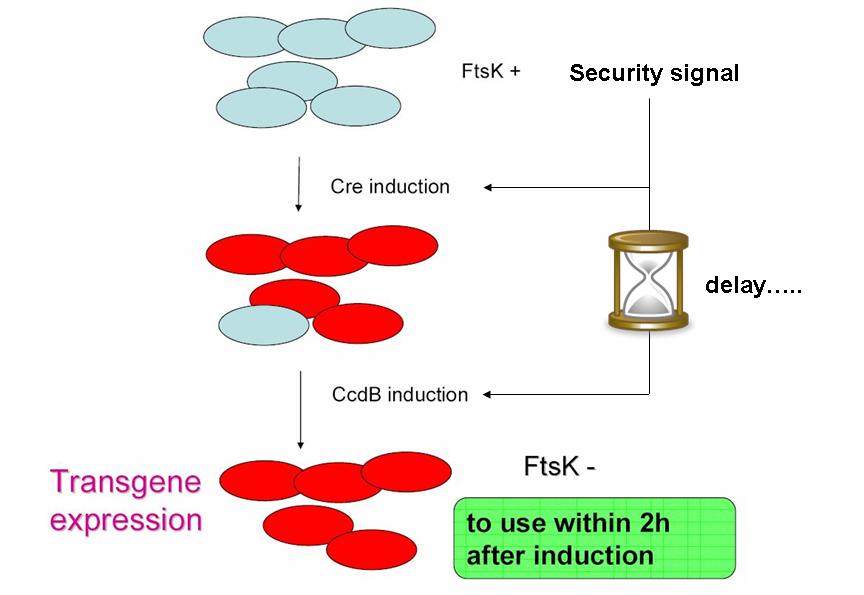
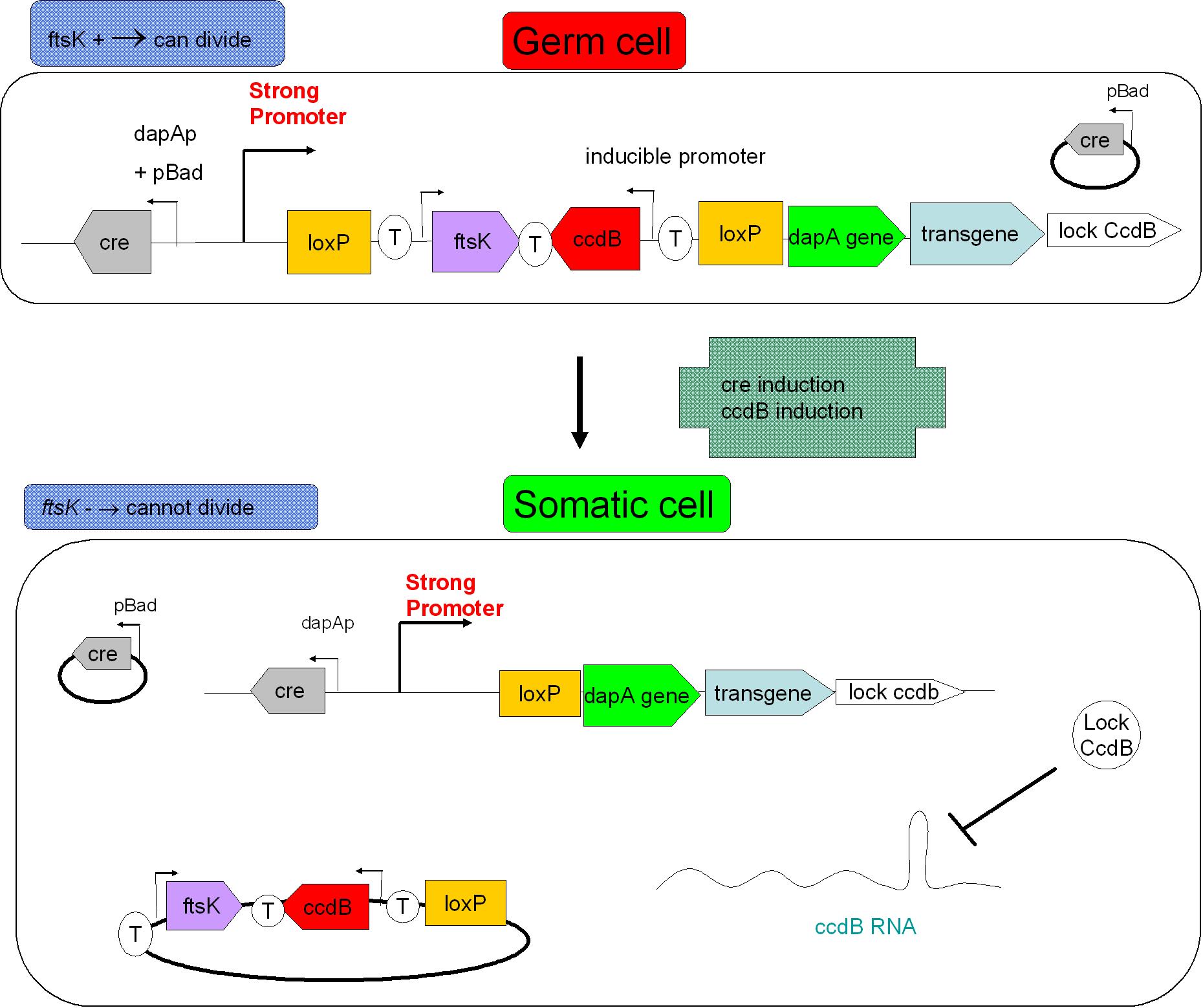
As indicated earlier, in order for the modified SMB to behave as a secure source of sterile somatic cells, two events should be initiated upon activation of a switch. This could be done in the following manner:
- EVENT 1
In order to achieve massive induction of recombination, a dormant copy of Cre site specific recombinase could be added to the SMB system. This copy would be plasmid born & would placed under control of an inducible promoter. But a certain proportion of G cells would most certainly persist undifferentiated.
- EVENT 2
The solution to second challenge could be coupling the high recombination switch to a delayed suicide switch in the gem line cells. include the expression of ccdB suicide gene in the germ line cells.
This could be the opportunity to practically apply one of the founding principles of synthetic biology: modularity in systems assembly & organisation. Indeed, a time delay circuit has already been described, constructed & theoretically studied (Sara Hooshangi et al. 2005, Sara Hooshangi et al. 2006 and Juan M. Pedraza et al. 2006). Using such a time delay circuit could allow using a single signal to induce event 1, followed after a delay by event 2.
In addition
Size based screening could be performed before environmental release. Indeed, S cells having lost ftsK expression present an interesting phenotype: they grow in size without division taking place, they "filament". The size threshold used in the cell sorting process should be determined by studying size distribution within germ cell and somatic cell populations. This could to improve security.
In the figure above, the recombination procress accompanying differentiation would lead to the transcription of a ccdB lock in somatic cells. This should protect the somatic cells from the suicide event that germ line cells go through. This lock could be a post transcriptional regulatory mechanism as this class of regulation requires less latency time before appearing.
References
Time delay circuit
- Ultrasensitivity and noise propagation in a synthetic transcriptional cascade
Sara Hooshangi, Stephan Thiberge, and Ron Weiss PNAS March 8, 2005 vol.102 no. 10 p3581–3586
- Noise Propagation in Gene Networks
Juan M. Pedraza and Alexander van Oudenaarden SCIENCE March 25, 2005 vol.307
- The effect of negative feedback on noise propagation in transcriptional gene networks
Sara Hooshangi and Ron Weiss CHAOS 16, 026108 (2006)
RNA based regulation in Synthetic Biology
- RNA synthetic biology.
Farren J Isaacs1, Daniel J Dwyer2,3 & James J Collins3. NATURE BIOTECHNOLOGY Review VOLUME 24 NUMBER 5 MAY 2006.
Engineered riboregulators
- Engineered riboregulators enable post-transcriptional control of.
Isaacs, F.J. et al. gene expression. Nat. Biotechnol. 22, 841–847 (2004).
Ribosome mRNA pairing
- network of orthogonal ribosome-mRNA pairs.
Rackham, O. & Chin, J.W. A. Nat.Chem. Biol. 1, 159–166 (2005).
- Cellular logic with orthogonal ribosomes.
Rackham, O. & Chin, J.W. J. Am. Chem. Soc. 127, 17584–17585 (2005).
Engineered ligand-controlled riboregulators (Riboswitches)
- Controlling gene expression in living cells through small
molecule-RNA interactions. Werstuck, G. & Green, M.R. Science 282, 296–298 (1998).
- theophylline responsive riboswitch based on helix slipping controls
gene expression in vivo.. Suess, B., Fink, B., Berens, C., Stentz, R. & Hillen, W. A. Nucleic Acids Res. 32, 1610–1614 (2004).
- Programmable ligand-controlled riboregulators of eukaryotic
gene expression. Bayer, T.S. & Smolke, C.D. Nat. Biotechnol. 23, 337–343 (2005).
Back to Perspectives