Paris/DesignProcess
From 2007.igem.org
Eismoustique (Talk | contribs) (→Overview of the project) |
David.bikard (Talk | contribs) (→Constraints on essential gene choice) |
||
| (37 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
== <center>SMB Design Process</center>== | == <center>SMB Design Process</center>== | ||
| - | + | The first question that needs to be addressed in the design process of our system is that of the nature of the dependency relationships between its cellular components. Namely, what cross-dependency relationship between the somatic & germ line cells can best ensure “ecological” equilibrium & “evolutionary” stability between the two cell types? | |
---- | ---- | ||
| - | ===The soma needs the | + | ===The soma needs the germline=== |
====What dependency relationship?==== | ====What dependency relationship?==== | ||
| - | A radical mean of ensuring somatic cell dependence on | + | A radical mean of ensuring somatic cell dependence on germline is to make somatic cells sterile. That is, if somatic cells are unable to replicate, then they exclusively depend on G=>S differentiation for their existence. Furthermore, in this way it is impossible for S cells to overtake the population. |
====How can sterility be achieved?==== | ====How can sterility be achieved?==== | ||
| - | A great number of essential genes have been identified since the early days of E.coli genetics. An essential gene is one for which a mutant strain cannot grow at 37° on rich medium (LB). These genes include the essential division genes. Mutants of | + | A great number of essential genes have been identified since the early days of E.coli genetics. An essential gene is one for which a mutant strain cannot grow at 37° on rich medium (LB). These genes include the essential division genes. Mutants of these genes cannot divide. Suppressing the expression of such a gene would thus make the cell sterile. |
=====Differentiation mechanism===== | =====Differentiation mechanism===== | ||
| - | As discussed above, differentiation can be epigenetic (changes in genes expression pattern) or genetic (change in DNA sequence). In order to suppress the expression of an essential gene, we could either place it under the control of an inducible promoter that we would repress (epigenetic differentiation), or remove the gene itself through recombination (genetic differentiation). A genetic mechanism of differentiation is | + | As discussed above, differentiation can be epigenetic (changes in genes expression pattern) or genetic (change in DNA sequence). In order to suppress the expression of an essential gene, we could either place it under the control of an inducible promoter that we would repress (epigenetic differentiation), or remove the gene itself through recombination (genetic differentiation). A genetic mechanism of differentiation is more robust than an epigenetic one in a way that the system artificially implemented in the cell would escape with more difficulties. |
| - | Therefore, we decided that the | + | Therefore, we decided that the Germline=>Soma differentiation mechanism should be genetic (as opposed to epigenetic). Sterility in S cells will be achieved by deleting an essential gene from the genome during the differentiation process. This will be done through site-specific recombination. |
=====Constraints on essential gene choice===== | =====Constraints on essential gene choice===== | ||
| Line 32: | Line 32: | ||
* '''Gene isolation:''' In our final construct, cassettes will be inserted before and after the essential gene. We want our gene to be isolated, so as not to disturb the expression of other nearby genes. It should not for instance be embedded in an operon. See here for more details about the construction method. | * '''Gene isolation:''' In our final construct, cassettes will be inserted before and after the essential gene. We want our gene to be isolated, so as not to disturb the expression of other nearby genes. It should not for instance be embedded in an operon. See here for more details about the construction method. | ||
| - | We have finally chosen ftsK as the gene to be used. It is an isolated gene. Also, ts (thermo sensitive) strains for this gene give filamentous cells | + | We have finally chosen ftsK as the gene to be used. It is an isolated gene. Also, ts (thermo sensitive) strains for this gene give filamentous cells at the restrictive temperature that continue growing for an equivalent of over 20 generations resulting in impressively long cells as in the following picture where DNA is colored in red. |
| - | + | ||
| + | [[Image:FtsK.png|200px|center]] | ||
---- | ---- | ||
| Line 57: | Line 57: | ||
* '''Feedback:''' For purposes described below, we would like to have a sensor device sensitive to our metabolite concentration. | * '''Feedback:''' For purposes described below, we would like to have a sensor device sensitive to our metabolite concentration. | ||
| - | We finally choose diaminopimelate (DAP) as our metabolite. The germ line will be deleted for the dapA gene, an essential gene in the peptidoglycan and lysine biosynthesis pathways. For details about our choice [[Paris/Auxotrophy|see here]]. | + | We finally choose diaminopimelate (DAP) as our metabolite. The germ line will be deleted for the dapA gene, an essential gene in the peptidoglycan and lysine biosynthesis pathways. For details about our choice [[Paris/Auxotrophy|'''see here''']]. |
| - | In order to confirm that our choice fits well the above mentioned criteria, we conducted several | + | |
| + | |||
| + | A particularly appealing feature of dapA for our system is that some bacterial species have DapA enzymes that are not submitted to allosteric inhibition by DAP. The activity of E.coli DapA protein is to some extent inhibited by DAP. However, Bacillus subtilis DapA enzyme is not submitted to such a control. This enables the possibility | ||
| + | of using B.subtilis dapA gene in our system for DAP overexpression. | ||
| + | |||
| + | |||
| + | In order to confirm that our choice fits well the above mentioned criteria, we conducted several experiments. (See the [[Paris/Results|results page]]) | ||
====How will auxotroph germ line cells differentiate into prototroph somatic cells?==== | ====How will auxotroph germ line cells differentiate into prototroph somatic cells?==== | ||
| Line 77: | Line 83: | ||
===What differentiation rate?=== | ===What differentiation rate?=== | ||
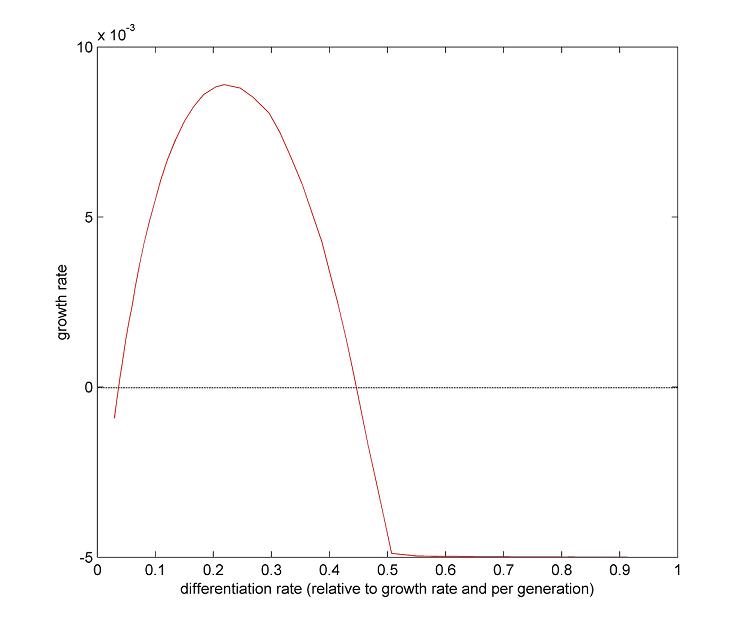
| - | [[Image:Paris_RecRate.jpg|right| | + | |
| + | [[Image:Paris_RecRate.jpg|right|350px]] | ||
| + | |||
| + | Our system's success relies on the differentiation rate of the germ line into soma. An intuitive reasoning shows that we cannot have more than 50% of differentiation per generation. Otherwise, the germ line will only decrease. But this is not the only constraint to take into account. | ||
For our germ line to grow fast, we need to maximize the DAP production. This means that we need to maximize the excretion of DAP by the soma and to maximize the proportion of the soma itself. The number of somatic cells is given by the differentiation rate of germ line cells and the life time of somatic cells. Thus we need to maximize the '''life time of somatic cells''' and the '''differentiation rate''' of the germ line. | For our germ line to grow fast, we need to maximize the DAP production. This means that we need to maximize the excretion of DAP by the soma and to maximize the proportion of the soma itself. The number of somatic cells is given by the differentiation rate of germ line cells and the life time of somatic cells. Thus we need to maximize the '''life time of somatic cells''' and the '''differentiation rate''' of the germ line. | ||
| Line 84: | Line 93: | ||
<br> | <br> | ||
[[Image:Tradeoff.jpg|left|400px]] | [[Image:Tradeoff.jpg|left|400px]] | ||
| - | |||
| - | We clearly see the conflict here, which will ultimately lead to a | + | [[Image:Paris_GrowthOnRec.jpg|right|400px]] |
| - | <br> | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| + | |||
| + | We clearly see the conflict here, which will ultimately lead to a trade-off between the germ line proportion and its generation time. As explained above, the differentiation rate cannot be over 50% per generation if we want our synthetic organism to grow. What this trade-off suggests is that there is an optimum differentiation rate somewhere between 0 and 50% recombinant per generation. The effect of the differentiation rate on the growth rate is displayed in [[Paris/Robustness_and_optimization|the modeling of our system]]. On the right you can see modeling results showing the relation between growth rate and recombination rate. | ||
| + | <br> | ||
| + | |||
====What recombination system?==== | ====What recombination system?==== | ||
| - | According to the previous point, we need a recombination system whose rate can be | + | According to the previous point, we need a recombination system whose rate can be controlled. Ideally we wish to obtain recombination at any chosen frequency. If this is impossible, we need at least the recombination rate to be below 50% per generation, but high enough for the germ line to be sufficiently fed. Another constraint on the recombination system, is that we want differentiation to be unidirectional. We thus chose the Cre/Lox recombination system, for the following reasons: |
| - | *it is readily available, largely used and well described | + | * it is readily available, largely used and well described. |
| - | *lox66 and lox71 recombination sites have been described to produce unidirectional recombination ''( Cre recombinase-mediated inversion using lox66 and lox71: method to introduce conditional point mutations into the CREB-binding protein, Zuwen Zhang and Beat Lutz, Nucleic Acids Research)'' | + | * lox66 and lox71 recombination sites have been described to produce unidirectional recombination ''( Cre recombinase-mediated inversion using lox66 and lox71: method to introduce conditional point mutations into the CREB-binding protein, Zuwen Zhang and Beat Lutz, Nucleic Acids Research)'' |
| Line 102: | Line 114: | ||
====Optimization through feedback==== | ====Optimization through feedback==== | ||
| - | It is clear that the average recombination rate must be between 0 and 50% per generation. Nevertheless, | + | It is clear that the average recombination rate must be between 0 and 50% per generation. Nevertheless, it might be possible to maximize growth rate by adjusting the recombination rate to DAP level. Germline differentiation rate could increase upon DAP starvation. The quantitative population analysis [[Paris/Robustness_and_optimization|model]] investigates this. In order to achieve the desired adaptive behavior, Cre expression must be adjusted to DAP concentration. This would be easily done if there where a promoter sensitive to DAP concentration, which seems to be the case of the dapA promoter (dapAp). We therefore cloned and characterized this promoter. |
| + | |||
| + | Two different system designs with regard to recombination frequency control are possible: | ||
| + | |||
| + | *A system with constant recombination frequency | ||
| + | *A system with adaptive recombination frequency | ||
| Line 109: | Line 126: | ||
===Overview of the project=== | ===Overview of the project=== | ||
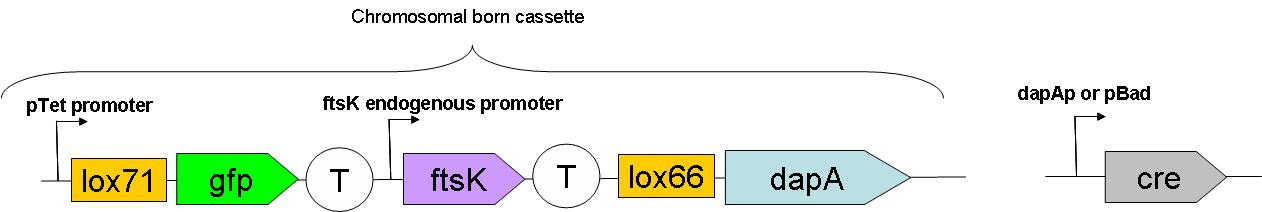
| - | + | After going through the system design process, we have selected a number of basic features to be included in the SMB system. A cassette, functional when inserted in the chromosome, needs to be generated. | |
| - | + | ||
| - | + | ||
| - | [[Image: | + | [[Image:Paris_FinalConstruct2.JPG|650px]] |
| - | This full construct | + | This full construct is in process of insertion into the genome. |
| - | As can be seen | + | As can be seen in this schematic representation of the SMB genomic backbone cassette, at basal genomic state (in Germ line cells, before recombination): |
*pTet promoter drives the expression of gfp. | *pTet promoter drives the expression of gfp. | ||
| Line 126: | Line 141: | ||
*dapA gene is in a dormant state since it lacks a promoter to drive its transcription. | *dapA gene is in a dormant state since it lacks a promoter to drive its transcription. | ||
| + | Regarding Cre expression control: | ||
| + | |||
| + | *Either Cre expression is fixed (under pBAd promoter control) | ||
| + | *Or it is submitted to DAP starvation feedback (under dapAp control). The potential impact of using a constitutively-expressed or regulated cre gene is discussed in detail in the [[Paris/Robustness_and_optimization|modelling section]]. | ||
| + | |||
| + | |||
| + | Regarding the dapA gene to be used: | ||
| + | |||
| + | *Either dapA from MG1655 E.coli is used. The protein product of this gene will be submitted to allosteric control by DAP | ||
| + | *Or B.subtilis dapA gene is used. The protein expressed will not suffer DAP inhibition. This could increase DAP production. The potential impact of maximizing DAP production by the soma on the robustness of SMB growth is discussed in the [[Paris/Robustness_and_optimization#Optimization|modelling section]]. | ||
| + | |||
| + | To learn more about the construction process, [[Paris/ConstructionProcess|click here]] or read through to the construction process section. | ||
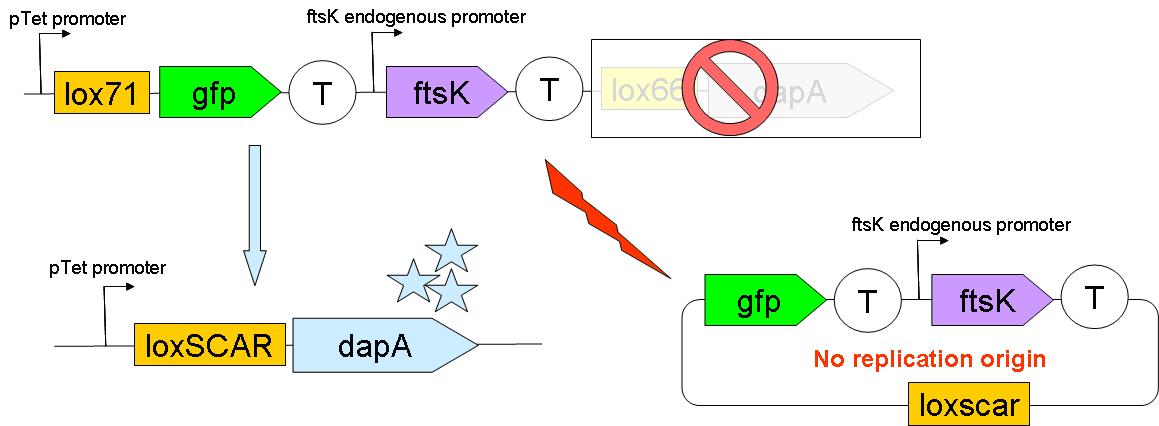
Upon G to S differentiation, the following genomic reassembly should take place: | Upon G to S differentiation, the following genomic reassembly should take place: | ||
| Line 135: | Line 162: | ||
Cre mediated lox recombination should lead to: | Cre mediated lox recombination should lead to: | ||
| - | *excision of ftsK gene from the genome | + | *excision of ftsK gene from the genome onto a circular non-replicative DNA molecule. This circular molecule may remain for quite long in the cell, and as the ftsK goes with its promoter, it may well still be expressed after exision. This would lead to a cell still able to divide but producing soma only. Nevertheless it seems that the division is highly dependent on FtsK concentration, and its presence in single copy might very well not be sufficient for division. (FtsK, a literate chromosome segregation machine. Bigot S et ''al.'', Mol Microbio) |
*placing dapA gene under the control of pTet promoter. This should lead to dapA expression, & hopefully to DAP synthesis. | *placing dapA gene under the control of pTet promoter. This should lead to dapA expression, & hopefully to DAP synthesis. | ||
| - | + | <center><html> | |
| - | + | <object classid="clsid:D27CDB6E-AE6D-11cf-96B8-444553540000" codebase="http://download.macromedia.com/pub/shockwave/cabs/flash/swflash.cab#version=6,0,29,0" width="700" height="860"> | |
| - | + | <param name="movie" value="http://biosynthetique.free.fr/videos/replic2.swf"> | |
| + | <param name="quality" value="high"> | ||
| + | <embed src="http://biosynthetique.free.fr/videos/replic2.swf" quality="high" pluginspage="http://www.macromedia.com/go/getflashplayer" type="application/x-shockwave-flash" width="700" height="860"></embed></object> | ||
| + | </html></center> | ||
---- | ---- | ||
| Line 167: | Line 197: | ||
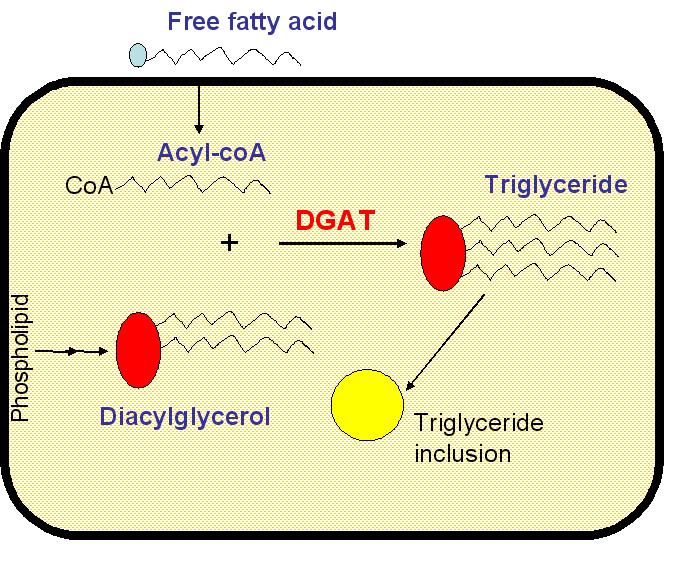
Triglycerides are not a natural product of E. coli metabolism as it lacks diacylglycerol acyl-transferase (DGAT). DGAT catalyses the reaction of glycerol esterifaication by fatty acids. However, all the compounds necessary for the triglyceride synthesis are present in the cytoplasm: diacylglycerol is an intermediate of phospholipid catabolism (source ecocyc.org) and free fatty acids are imported from extracellular medium. Wild-type E. coli strains can indeed grow on free fatty acid medium (oleate for example). E. Coli has a Long Chain Fatty Acid (LCFA) transporter, FadL. Another protein: FadR, a long chain acyl-CoA-responsive transcription factor, controls the expression of nine genes primarily involved in fatty acid degradation and biosynthesis (FadL is thus induced by FadR) and β-oxidation enzymes.<br> <br> | Triglycerides are not a natural product of E. coli metabolism as it lacks diacylglycerol acyl-transferase (DGAT). DGAT catalyses the reaction of glycerol esterifaication by fatty acids. However, all the compounds necessary for the triglyceride synthesis are present in the cytoplasm: diacylglycerol is an intermediate of phospholipid catabolism (source ecocyc.org) and free fatty acids are imported from extracellular medium. Wild-type E. coli strains can indeed grow on free fatty acid medium (oleate for example). E. Coli has a Long Chain Fatty Acid (LCFA) transporter, FadL. Another protein: FadR, a long chain acyl-CoA-responsive transcription factor, controls the expression of nine genes primarily involved in fatty acid degradation and biosynthesis (FadL is thus induced by FadR) and β-oxidation enzymes.<br> <br> | ||
| - | + | <br><br> | |
| - | + | ||
| Line 176: | Line 205: | ||
*'''Which enzyme?''' | *'''Which enzyme?''' | ||
| - | We decided to use DGAT enzyme imported from a bacterium closely related to E. coli for metabolic compatibility | + | We decided to use DGAT enzyme imported from a bacterium closely related to E. coli for metabolic compatibility. Acinetobacter calcoaceticus ADP1, a Gram negative bacillus was a good candidate. Its DGAT enzyme is also an acyl-CoA fatty alcohol acyltransferase (wax ester synthase) and catalyzes the final condensation of acyl-CoA and fatty alcohol. But knowing that E.coli does not produce fatty alcohol, this reaction is probably not avaible. (for more information see <bbpart>BBa_I718002</bbpart>).<br> |
Latest revision as of 03:26, 27 October 2007
Contents |
SMB Design Process
The first question that needs to be addressed in the design process of our system is that of the nature of the dependency relationships between its cellular components. Namely, what cross-dependency relationship between the somatic & germ line cells can best ensure “ecological” equilibrium & “evolutionary” stability between the two cell types?
The soma needs the germline
What dependency relationship?
A radical mean of ensuring somatic cell dependence on germline is to make somatic cells sterile. That is, if somatic cells are unable to replicate, then they exclusively depend on G=>S differentiation for their existence. Furthermore, in this way it is impossible for S cells to overtake the population.
How can sterility be achieved?
A great number of essential genes have been identified since the early days of E.coli genetics. An essential gene is one for which a mutant strain cannot grow at 37° on rich medium (LB). These genes include the essential division genes. Mutants of these genes cannot divide. Suppressing the expression of such a gene would thus make the cell sterile.
Differentiation mechanism
As discussed above, differentiation can be epigenetic (changes in genes expression pattern) or genetic (change in DNA sequence). In order to suppress the expression of an essential gene, we could either place it under the control of an inducible promoter that we would repress (epigenetic differentiation), or remove the gene itself through recombination (genetic differentiation). A genetic mechanism of differentiation is more robust than an epigenetic one in a way that the system artificially implemented in the cell would escape with more difficulties.
Therefore, we decided that the Germline=>Soma differentiation mechanism should be genetic (as opposed to epigenetic). Sterility in S cells will be achieved by deleting an essential gene from the genome during the differentiation process. This will be done through site-specific recombination.
Constraints on essential gene choice
Remains the question of the choice of the essential gene to be used. We have decided to base our choice on two criteria:
- Longevity of the S cells: S cells even if unable to divide should be able to live as long as possible. (an explanation for this is given below)
- Gene isolation: In our final construct, cassettes will be inserted before and after the essential gene. We want our gene to be isolated, so as not to disturb the expression of other nearby genes. It should not for instance be embedded in an operon. See here for more details about the construction method.
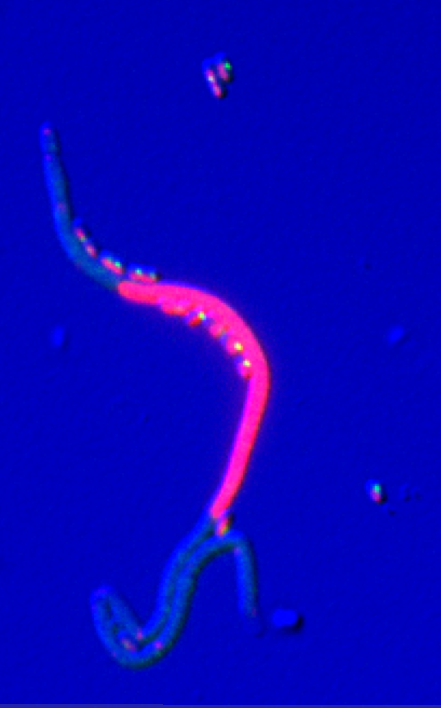
We have finally chosen ftsK as the gene to be used. It is an isolated gene. Also, ts (thermo sensitive) strains for this gene give filamentous cells at the restrictive temperature that continue growing for an equivalent of over 20 generations resulting in impressively long cells as in the following picture where DNA is colored in red.
The germ line needs the soma
What dependency relationship?
In our synthetic organism, the germ line is auxtroph for a given nutrient that is provided by the soma. In this way, a part of the germ line needs to differentiate, in order to feed the undifferentiated fraction.
Why an auxotrophy dependence?
Previous works have shown that two types of cell, each auxotroph for a different metabolite, can rescue each other when grown in the same minimum media. This means that some auxotrophy should be rescued in a coculture with prototroph cells. Plus, it is very easy to do!
What auxotrophy?
In order to choose our auxotrophy, we need to take several constraints into account:
- No simple bypass or reversion: The auxotrophy phenotype of the germ line must be stable. There must be no simple metabolic bypass or reversion possibilities.
- Overproduction and excretion: Prototroph cells must be able to overproduce and excrete the metabolite, or at least to release it when dying.
- Absorption: auxotroph cells should be able to live in very low concentration of this metabolite. Indeed we should expect only little metabolite to be excreted.
- Survival: auxotroph cells should be able to live as long as possible when deprived from the metabolite.
- No growth on LB: We would like our synthetic organism to be able to grow on rich medium. This would facilitate our lab work, and more generally allow our synthetic organism to grow in a variety of conditions.
- Feedback: For purposes described below, we would like to have a sensor device sensitive to our metabolite concentration.
We finally choose diaminopimelate (DAP) as our metabolite. The germ line will be deleted for the dapA gene, an essential gene in the peptidoglycan and lysine biosynthesis pathways. For details about our choice see here.
A particularly appealing feature of dapA for our system is that some bacterial species have DapA enzymes that are not submitted to allosteric inhibition by DAP. The activity of E.coli DapA protein is to some extent inhibited by DAP. However, Bacillus subtilis DapA enzyme is not submitted to such a control. This enables the possibility of using B.subtilis dapA gene in our system for DAP overexpression.
In order to confirm that our choice fits well the above mentioned criteria, we conducted several experiments. (See the results page)
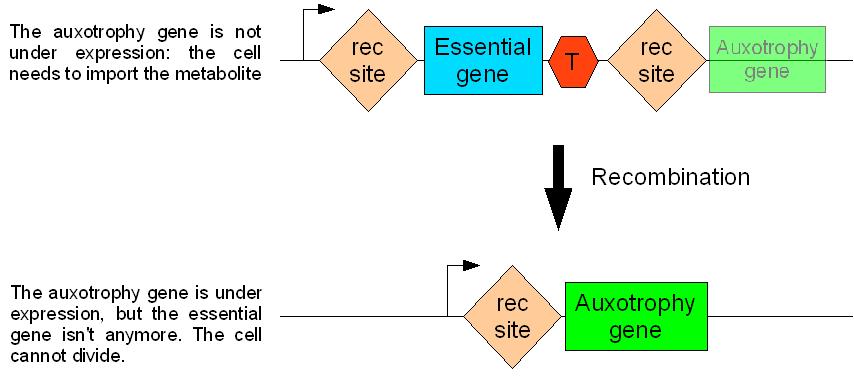
How will auxotroph germ line cells differentiate into prototroph somatic cells?
We found an elegant way to solve this problem. This is best described by the following schema.
rec is a site specific recombination sequence
T is a strong terminator
System scale specifications
What differentiation rate?
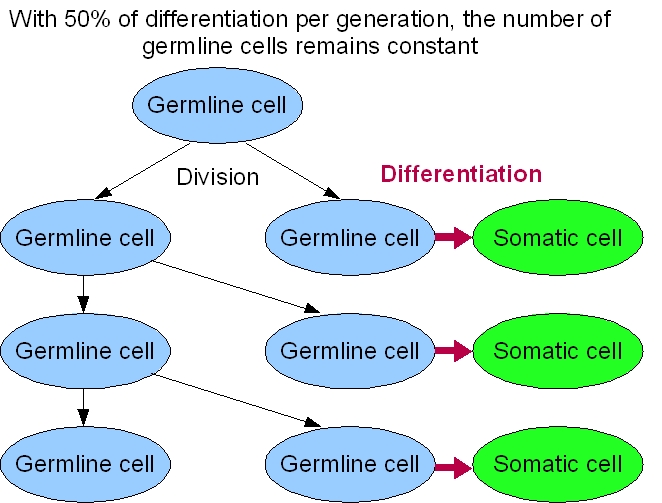
Our system's success relies on the differentiation rate of the germ line into soma. An intuitive reasoning shows that we cannot have more than 50% of differentiation per generation. Otherwise, the germ line will only decrease. But this is not the only constraint to take into account.
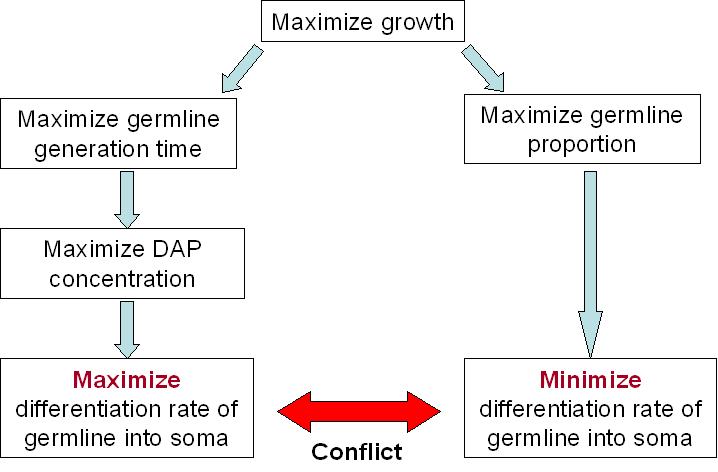
For our germ line to grow fast, we need to maximize the DAP production. This means that we need to maximize the excretion of DAP by the soma and to maximize the proportion of the soma itself. The number of somatic cells is given by the differentiation rate of germ line cells and the life time of somatic cells. Thus we need to maximize the life time of somatic cells and the differentiation rate of the germ line. On the other hand, we want our synthetic organism to grow as fast as possible. And it will grow faster if the proportion of germ line cells is bigger, since somatic cells do not divide. This means that we have to minimize the differentiation rate.
We clearly see the conflict here, which will ultimately lead to a trade-off between the germ line proportion and its generation time. As explained above, the differentiation rate cannot be over 50% per generation if we want our synthetic organism to grow. What this trade-off suggests is that there is an optimum differentiation rate somewhere between 0 and 50% recombinant per generation. The effect of the differentiation rate on the growth rate is displayed in the modeling of our system. On the right you can see modeling results showing the relation between growth rate and recombination rate.
What recombination system?
According to the previous point, we need a recombination system whose rate can be controlled. Ideally we wish to obtain recombination at any chosen frequency. If this is impossible, we need at least the recombination rate to be below 50% per generation, but high enough for the germ line to be sufficiently fed. Another constraint on the recombination system, is that we want differentiation to be unidirectional. We thus chose the Cre/Lox recombination system, for the following reasons:
- it is readily available, largely used and well described.
- lox66 and lox71 recombination sites have been described to produce unidirectional recombination ( Cre recombinase-mediated inversion using lox66 and lox71: method to introduce conditional point mutations into the CREB-binding protein, Zuwen Zhang and Beat Lutz, Nucleic Acids Research)
We want to control the recombination frequency by adjusting the expression level of the Cre recombinase. To do this, we cloned the Cre recombinase under the control of the pBad promoter (inducible through arabinose). We also decided to clone our Cre production device (araC-pBad>>rbs-Cre) on a low copy number plasmid, to broaden the reachable expression rates.
Recombinaison frequency measurment experiments
Optimization through feedback
It is clear that the average recombination rate must be between 0 and 50% per generation. Nevertheless, it might be possible to maximize growth rate by adjusting the recombination rate to DAP level. Germline differentiation rate could increase upon DAP starvation. The quantitative population analysis model investigates this. In order to achieve the desired adaptive behavior, Cre expression must be adjusted to DAP concentration. This would be easily done if there where a promoter sensitive to DAP concentration, which seems to be the case of the dapA promoter (dapAp). We therefore cloned and characterized this promoter.
Two different system designs with regard to recombination frequency control are possible:
- A system with constant recombination frequency
- A system with adaptive recombination frequency
Overview of the project
After going through the system design process, we have selected a number of basic features to be included in the SMB system. A cassette, functional when inserted in the chromosome, needs to be generated.
This full construct is in process of insertion into the genome.
As can be seen in this schematic representation of the SMB genomic backbone cassette, at basal genomic state (in Germ line cells, before recombination):
- pTet promoter drives the expression of gfp.
- The expression of ftsK is controlled by its natural promoter.
- ftsK is isolated from pTet promoter by the intercalation of Terminator (B0015 terminator).
- dapA gene is in a dormant state since it lacks a promoter to drive its transcription.
Regarding Cre expression control:
- Either Cre expression is fixed (under pBAd promoter control)
- Or it is submitted to DAP starvation feedback (under dapAp control). The potential impact of using a constitutively-expressed or regulated cre gene is discussed in detail in the modelling section.
Regarding the dapA gene to be used:
- Either dapA from MG1655 E.coli is used. The protein product of this gene will be submitted to allosteric control by DAP
- Or B.subtilis dapA gene is used. The protein expressed will not suffer DAP inhibition. This could increase DAP production. The potential impact of maximizing DAP production by the soma on the robustness of SMB growth is discussed in the modelling section.
To learn more about the construction process, click here or read through to the construction process section.
Upon G to S differentiation, the following genomic reassembly should take place:
Cre mediated lox recombination should lead to:
- excision of ftsK gene from the genome onto a circular non-replicative DNA molecule. This circular molecule may remain for quite long in the cell, and as the ftsK goes with its promoter, it may well still be expressed after exision. This would lead to a cell still able to divide but producing soma only. Nevertheless it seems that the division is highly dependent on FtsK concentration, and its presence in single copy might very well not be sufficient for division. (FtsK, a literate chromosome segregation machine. Bigot S et al., Mol Microbio)
- placing dapA gene under the control of pTet promoter. This should lead to dapA expression, & hopefully to DAP synthesis.
E. Colight: towards a new slim diet
Project
Triglycerides are composed of a molecule of glycerol esterified by three fatty acids. When ingested, triglycerides are hydrolysed by lipases, in the stomach and the duodenum, into glycerol and free fatty acids. Enterocytes are can only absorb free fatty acids and glycerol. These are subsequently recombined in the cytoplasm into triglycerides. Triglycerides are then freed in the lymphatic system and then in the blood within fatty vesicles called chylomicrons.
When lipid uptake versus energy consumption and loss is unbalanced, we accumulate fat. Knowing that gut is full with bacteria forming the gut microflora (we have 1013 cells in our body and 1014 bacteria with the majority in the gut!), we envision engineering bacteria capable of absorbing fatty acids and storing them in the form of triglycerides intracellular inclusion. These triglycerides would not be absorbed by enterocytes! Lipid input would decrease! Eat fat, don't get fat!
In fact, a drug, orlistat, already exists and shows decreasing lipid input works in obesity. It comes from a bacterial lipase inhibitor (from Streptomyces toxytricini). It inhibits pancreatic lipase and is used to cure obese people and type 2 diabetes with hypocaloric cure. At the standard prescription dose of 120 mg three times daily before meals, orlistat prevents approximately 30% of dietary fat from being absorbed (Thomson PDR, 2006).
E.coli is the most used bacterium in synthetic biology and... belongs to the gut microflora! We wish to genetically engineer E. coli into Ecolight to store triglycerides into inclusions! Knowing that 40% of E.coli is renewed every day, these triglyceride-filled bacteria will leave the gut with faeces!
The result of this work should, in a second phase of our work, be combined with the “security device” derived from the SMB described above. This is further discussed in PERSPECTIVES.
Technical details
As a perspective for our SMB, we want to show that it can be a tool for metabolic engineering (see perspectives). We started to develop the idea of making E. Coli store fatty acids in the form of triglycerides. Such an engineered bacteria could be ingested to absorb fatty acids thus limiting the amount of fatty acids absorbed by the user! Eat fat don't get fat !
- Why should it work?
Triglycerides are not a natural product of E. coli metabolism as it lacks diacylglycerol acyl-transferase (DGAT). DGAT catalyses the reaction of glycerol esterifaication by fatty acids. However, all the compounds necessary for the triglyceride synthesis are present in the cytoplasm: diacylglycerol is an intermediate of phospholipid catabolism (source ecocyc.org) and free fatty acids are imported from extracellular medium. Wild-type E. coli strains can indeed grow on free fatty acid medium (oleate for example). E. Coli has a Long Chain Fatty Acid (LCFA) transporter, FadL. Another protein: FadR, a long chain acyl-CoA-responsive transcription factor, controls the expression of nine genes primarily involved in fatty acid degradation and biosynthesis (FadL is thus induced by FadR) and β-oxidation enzymes.
- Which enzyme?
We decided to use DGAT enzyme imported from a bacterium closely related to E. coli for metabolic compatibility. Acinetobacter calcoaceticus ADP1, a Gram negative bacillus was a good candidate. Its DGAT enzyme is also an acyl-CoA fatty alcohol acyltransferase (wax ester synthase) and catalyzes the final condensation of acyl-CoA and fatty alcohol. But knowing that E.coli does not produce fatty alcohol, this reaction is probably not avaible. (for more information see BBa_I718002).