Peking The Projects
From 2007.igem.org
Zheng Qinsi (Talk | contribs) (→Towards Self-differentiated Bacterial Assembly Line) |
|||
| (44 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| - | + | <div style="background-color:#f7fbd7;"> | |
| - | {| style="color:#000000; background-color:# | + | [[Image:Peking_Final_logo.jpg|310px]][[Image:Peking Head banner-3.jpg|570px]] |
| + | {| style="color:#000000; background-color:#EEf8b3;" cellpadding="5" cellspacing="1" border="0" bordercolor="#1100ff" width="933" | ||
![[Peking|Peking]] | ![[Peking|Peking]] | ||
![[Peking The Projects|The Projects]] | ![[Peking The Projects|The Projects]] | ||
![[Peking The Community|The Team]] | ![[Peking The Community|The Team]] | ||
![[Peking The Fun|Misc & Fun]] | ![[Peking The Fun|Misc & Fun]] | ||
| - | ![http://openwetware.org/wiki/IGEM:Peking/2007 | + | ![http://www.openwetware.org/wiki/IGEM:Peking/2007 Our OWW] |
| - | ![[Peking Thanks| | + | ![[Peking Thanks|Acknowledgement]] |
|} | |} | ||
| - | =Self-differentiated Bacterial Assembly Line= | + | =Towards Self-differentiated Bacterial Assembly Line= |
| - | Our projects | + | Our projects concern with the ability for bacterial cells to '''differentiate''' out of homogeneous conditions into populations with the '''division of labor'''. We aim at devices conferring host cells with the ability to form cooperating groups spontaneously and to take consecutive steps sequentially even when the genetic background and environmental inputs are identical. To break the mirror in such homogeneous condition, we need two devices respectively responsible for '''temporal''' and '''spatial''' differentiation. The implementation and application of such devices will lead to a picture of bioengineering where complex programs consisted of sequential steps (structure oriented programs) and cooperating agencies (forked instances of a single class, object and event oriented) can be embedded in a single genome. |
| - | + | ||
| + | <br> | ||
| + | Both the functions and design principles of the two devices are described in the below sections, the functions are presented by two figurative .gif animations to present what they do and how they make cells differentiate in the dimension of space and time. '''Detailed design and Experimental results are presented in two separate pages linked below'''. | ||
| - | ==[[Peking_Hop-Count|Hop Count with Conjugation]]== | + | <br> |
| + | About the name of projects, the two devices are named after their respective functions, but they are common for breaking symmetry, which is interesting and ubiquitous in science. Although this "differentiation" process resemble the development of multicellular organism, we tend to use a more bioengineering style analogy: assembly line. Or maybe after some years from now, this will not be just an analogy. | ||
| + | |||
| + | <table border="0" cellpadding="20" width=95% style="background-color:#f7fbd7;"> | ||
| + | <tr> | ||
| + | <th><div style="font: normal 1.2em georgia, serif">[[Peking_Hop-Count|'''Hop Count with Conjugation''']]<br></div> | ||
| + | ---- | ||
| + | Click [[Peking_Hop-Count|'''HERE''']] or visit [http://www.openwetware.org/wiki/IGEM:Peking/2007 '''OUR OWW site'''] for all the details of this part! | ||
| + | </th> | ||
| + | <th><div style="font: normal 1.2em georgia, serif">[[Peking_Push-on-push-off|'''Push-on-push-off Switch''']]<br></div> | ||
| + | ---- | ||
| + | Click [[Peking_Push-on-push-off| '''HERE''']] or visit [http://www.openwetware.org/wiki/IGEM:Peking/2007 '''OUR OWW site'''] for all the details of this part! | ||
| + | </th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td align="center">[[Image:Peking_Count_Function.gif]]</td> | ||
| + | <td align="center">[[Image:Peking_Switch_Function.gif]]</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
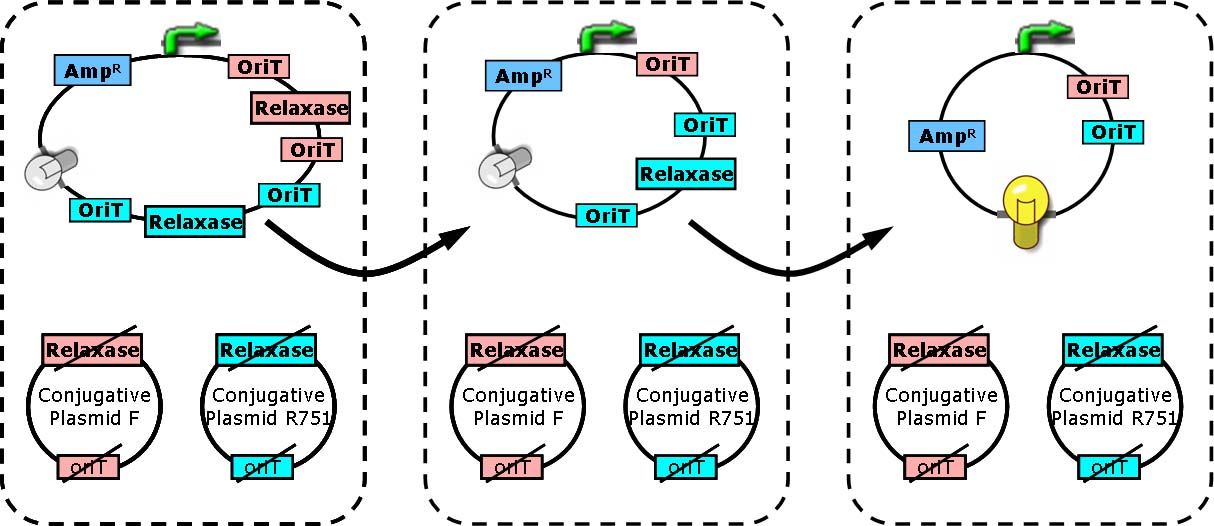
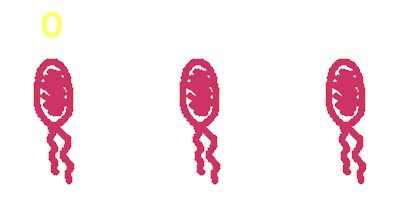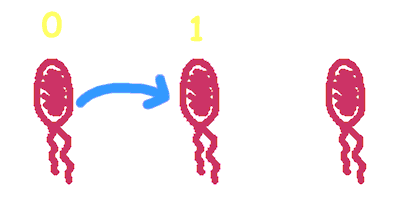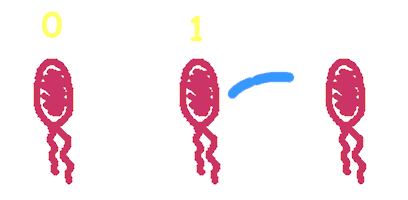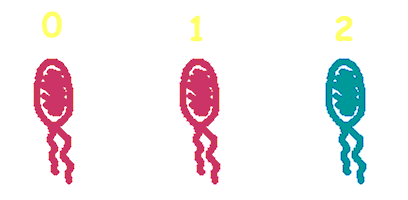
Hop counting is a cell-cell communication system in which the signals transfered among the cells record the number of times of transfer. With this system, cells can use the transmitted signal as topological cues of their neighborhood, and start the first step of differentiation and cooperation. | Hop counting is a cell-cell communication system in which the signals transfered among the cells record the number of times of transfer. With this system, cells can use the transmitted signal as topological cues of their neighborhood, and start the first step of differentiation and cooperation. | ||
We have adapted the natural conjugation systems to a communication device where the conjugative plasmids as the vector for the transfered signal. The numbering signals are directly encoded on the transmitted DNA sequence. | We have adapted the natural conjugation systems to a communication device where the conjugative plasmids as the vector for the transfered signal. The numbering signals are directly encoded on the transmitted DNA sequence. | ||
| - | Utilizing a special character of the rolling-cycle replication of DNA during conjugation, we use a design where independent tandem oriT (<bbpart>BBa_J01002</bbpart> and more) pairs are inserted into the same plasmids, which lost one tandem region at one conjugation event. Thus the number of conjugation are recorded on the presence/absence of regions on the plasmid of arbitrary length . | + | Utilizing a special character of the rolling-cycle replication of DNA during conjugation, we use a design where independent tandem oriT (<bbpart>BBa_J01002</bbpart> and more) pairs are inserted into the same plasmids, which lost one tandem region at one conjugation event. Thus the number of conjugation are recorded on the presence/absence of regions on the plasmid of arbitrary length. |
| - | Sequential conjugation and deletion of the different oriT pairs can be achieved by fine control of the expression of the | + | Sequential conjugation and deletion of the different oriT pairs can be achieved by fine control of the expression of the trans-acting factors of the independent conjugation systems. In fact, we inserted the enzymes directly responsible for the initiation of conjugative DNA replication (the relaxase) followed by a terminator between the corresponding oriT. The expression of all the enzymes are under control of a unified promoter. The terminators ensure only the relaxase of the most upstream undeleted tandem oriT regions are expressed. This design guarantees the independent conjugative systems are deleted at a specified order. |
| + | |||
| + | [[Image:Tandem_conjugation.jpg|400px]] | ||
| + | |||
| + | Currently, we have tested and constructed three sets of independent conjugative systems and validated the feasibility of our design with the F system. The next step is to do the 3-step conjugation-counting process in a single strain. | ||
| + | |||
| + | </td> | ||
| + | <td valign="top"> | ||
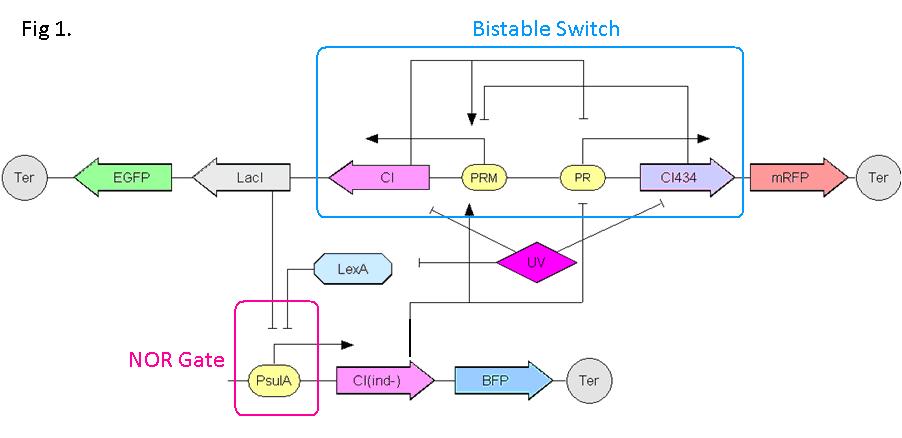
| + | Push-on-push-off switches are binary switches flipping to different direction under the same impulses, depending on the previous states it resides. It differed from a toggle switch that the forces flip the switch back and force are exactly the same. This allows the cells to do sequential logic utilizing both the information of present and pst events. | ||
| + | |||
| + | We have designed a gene regulatory circuit that senses Ultra-Violet light (UV) irradiation as input signal. Under intermittent irradiation, two reporter genes are to be induced alternately. The circuit can be divided into two associated parts: '''a bistable switch''' and a '''NOR gate'''. The bistable switch resembles a toggle switch where the NOR gate is controlled by the bistable switch and the input signal. On the other hand, the output of the NOR gate can switch the bistable state from one to the other. In this way, the NOR gate combined the information of present inputs and the previous states of the system, and thus flipping the switch in different directions based on the previous events, even though the present inputs are always the same. | ||
| - | + | [[Image:Peking Swich fig.1.JPG|400px]] | |
| - | + | Thus far, we have constructed and tested a series of NOR gates and one bistable switch, successfully obtaining the desired dynamics for the previous design(these dynamics may also be very useful to many other applications). The next is to combine the two together and tune the system to obtain the desired function. | |
| - | + | </td> | |
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
Latest revision as of 02:07, 27 October 2007
| Peking | The Projects | The Team | Misc & Fun | [http://www.openwetware.org/wiki/IGEM:Peking/2007 Our OWW] | Acknowledgement |
|---|
Towards Self-differentiated Bacterial Assembly Line
Our projects concern with the ability for bacterial cells to differentiate out of homogeneous conditions into populations with the division of labor. We aim at devices conferring host cells with the ability to form cooperating groups spontaneously and to take consecutive steps sequentially even when the genetic background and environmental inputs are identical. To break the mirror in such homogeneous condition, we need two devices respectively responsible for temporal and spatial differentiation. The implementation and application of such devices will lead to a picture of bioengineering where complex programs consisted of sequential steps (structure oriented programs) and cooperating agencies (forked instances of a single class, object and event oriented) can be embedded in a single genome.
Both the functions and design principles of the two devices are described in the below sections, the functions are presented by two figurative .gif animations to present what they do and how they make cells differentiate in the dimension of space and time. Detailed design and Experimental results are presented in two separate pages linked below.
About the name of projects, the two devices are named after their respective functions, but they are common for breaking symmetry, which is interesting and ubiquitous in science. Although this "differentiation" process resemble the development of multicellular organism, we tend to use a more bioengineering style analogy: assembly line. Or maybe after some years from now, this will not be just an analogy.
|
Click HERE or visit [http://www.openwetware.org/wiki/IGEM:Peking/2007 OUR OWW site] for all the details of this part! |
Click HERE or visit [http://www.openwetware.org/wiki/IGEM:Peking/2007 OUR OWW site] for all the details of this part! |
|---|---|
 |
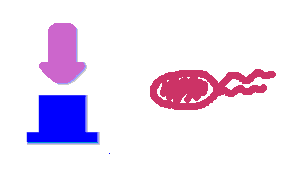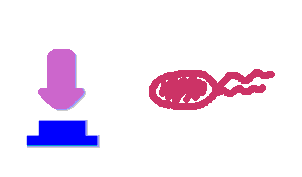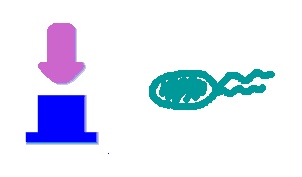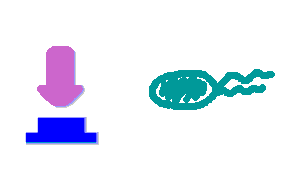 |
|
Hop counting is a cell-cell communication system in which the signals transfered among the cells record the number of times of transfer. With this system, cells can use the transmitted signal as topological cues of their neighborhood, and start the first step of differentiation and cooperation. We have adapted the natural conjugation systems to a communication device where the conjugative plasmids as the vector for the transfered signal. The numbering signals are directly encoded on the transmitted DNA sequence. Utilizing a special character of the rolling-cycle replication of DNA during conjugation, we use a design where independent tandem oriT (BBa_J01002 and more) pairs are inserted into the same plasmids, which lost one tandem region at one conjugation event. Thus the number of conjugation are recorded on the presence/absence of regions on the plasmid of arbitrary length. Sequential conjugation and deletion of the different oriT pairs can be achieved by fine control of the expression of the trans-acting factors of the independent conjugation systems. In fact, we inserted the enzymes directly responsible for the initiation of conjugative DNA replication (the relaxase) followed by a terminator between the corresponding oriT. The expression of all the enzymes are under control of a unified promoter. The terminators ensure only the relaxase of the most upstream undeleted tandem oriT regions are expressed. This design guarantees the independent conjugative systems are deleted at a specified order. Currently, we have tested and constructed three sets of independent conjugative systems and validated the feasibility of our design with the F system. The next step is to do the 3-step conjugation-counting process in a single strain. |
Push-on-push-off switches are binary switches flipping to different direction under the same impulses, depending on the previous states it resides. It differed from a toggle switch that the forces flip the switch back and force are exactly the same. This allows the cells to do sequential logic utilizing both the information of present and pst events. We have designed a gene regulatory circuit that senses Ultra-Violet light (UV) irradiation as input signal. Under intermittent irradiation, two reporter genes are to be induced alternately. The circuit can be divided into two associated parts: a bistable switch and a NOR gate. The bistable switch resembles a toggle switch where the NOR gate is controlled by the bistable switch and the input signal. On the other hand, the output of the NOR gate can switch the bistable state from one to the other. In this way, the NOR gate combined the information of present inputs and the previous states of the system, and thus flipping the switch in different directions based on the previous events, even though the present inputs are always the same. Thus far, we have constructed and tested a series of NOR gates and one bistable switch, successfully obtaining the desired dynamics for the previous design(these dynamics may also be very useful to many other applications). The next is to combine the two together and tune the system to obtain the desired function. |