Peking Hop-Count
From 2007.igem.org
Qu Mingzhi (Talk | contribs) m (→Measure conjugation efficiency of strain with conjugative plasmid F) |
Zheng Qinsi (Talk | contribs) (→'''Achievement''') |
||
| (18 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
![[Peking The Community|The Team]] | ![[Peking The Community|The Team]] | ||
![[Peking The Fun|Misc & Fun]] | ![[Peking The Fun|Misc & Fun]] | ||
| - | ![[Peking Thanks| | + | ![http://www.openwetware.org/wiki/IGEM:Peking/2007 Our OWW] |
| + | ![[Peking Thanks|Acknowledgement]] | ||
|} | |} | ||
| Line 14: | Line 15: | ||
[[Image:Peking Pass.JPG|60px]]'''Measure conjugation efficiency of multiple conjugative strains''' | [[Image:Peking Pass.JPG|60px]]'''Measure conjugation efficiency of multiple conjugative strains''' | ||
| - | [[Image:Peking Pass.JPG|60px]]'''Construct conjugative mutant strains with oriT and relaxase deleted''' | + | [[Image:Peking Pass.JPG|60px]]'''Construct conjugative mutant strains with oriT and relaxase deleted (and they WORK!)''' |
[[Image:Peking Pass.JPG|60px]]'''Measure efficiency and specificity of multiple riboregulator pairs''' | [[Image:Peking Pass.JPG|60px]]'''Measure efficiency and specificity of multiple riboregulator pairs''' | ||
| Line 72: | Line 73: | ||
Right: Control strain with no GFP expressed. | Right: Control strain with no GFP expressed. | ||
| - | Conclusion: | + | '''Conclusion:''' |
1. Genes express properly in a signaling plasmid with the oriT pair deleted. | 1. Genes express properly in a signaling plasmid with the oriT pair deleted. | ||
2. We successfully implemented and verified conjugation-deletion events with F system (binary hop count). | 2. We successfully implemented and verified conjugation-deletion events with F system (binary hop count). | ||
| + | |||
| + | ---- | ||
===Construct conjugative mutant strains with oriT and relaxase deleted=== | ===Construct conjugative mutant strains with oriT and relaxase deleted=== | ||
| - | Method 1: pKO3 system | + | '''Method 1: pKO3 system''' |
1. Use plasmid pKO3 to generate precise deletions of oriT region in conjugative plasmids through homologous recombination. | 1. Use plasmid pKO3 to generate precise deletions of oriT region in conjugative plasmids through homologous recombination. | ||
| Line 85: | Line 88: | ||
2. For working principle of pKO3 please refer to: Link A. et al., (1997) J Bacteriol 179: 6228. | 2. For working principle of pKO3 please refer to: Link A. et al., (1997) J Bacteriol 179: 6228. | ||
| - | 3. For more experimental details please visit [http://www.openwetware.org/wiki/IGEM:Peking/2007/Count:Knockout | + | 3. For more experimental details please visit [http://www.openwetware.org/wiki/IGEM:Peking/2007/Count:Knockout Knockout protocol in our OWW site]. |
| - | Method 2: Lambda Red system | + | '''Method 2: Lambda Red system''' |
1. Use plasmid pKD46 and homologous PCR products to inactivate genes of relaxase in conjugative plasmids. | 1. Use plasmid pKD46 and homologous PCR products to inactivate genes of relaxase in conjugative plasmids. | ||
| Line 97: | Line 100: | ||
| - | Method 3: Reversed cyclic PCR | + | '''Method 3: Reversed cyclic PCR''' |
1. Used for in vitro construction of plasmid pSC101, a conjugative plasmid in size of 9 kbp. | 1. Used for in vitro construction of plasmid pSC101, a conjugative plasmid in size of 9 kbp. | ||
| Line 104: | Line 107: | ||
| - | Achievement: | + | '''Achievement:''' |
| - | 1. Successfully deleted | + | 1. Successfully deleted genes of relaxase in plasmid F(<bbpart>BBa_I714035</bbpart>) , R751(<bbpart>BBa_I714037</bbpart>) and pSC101(<bbpart>BBa_I714038</bbpart>) . |
| - | + | ||
| - | + | ||
| + | 2. <bbpart>BBa_I714035</bbpart> successfully inactivated the conjugation process. <bbpart>BBa_I714037</bbpart> has a leaky of about 1/10. | ||
| + | ---- | ||
===Measure efficiency and specificity of multiple riboregulator pairs=== | ===Measure efficiency and specificity of multiple riboregulator pairs=== | ||
| - | In order to provide an interface for interaction between our conjugation system and general genetic devices to realize the goal of assembly line, a standardized way of controlling target gene expression is necessary. Here we mainly concern the application of riboregulators, which was successfully introduced by [[ | + | In order to provide an interface for interaction between our conjugation system and general genetic devices to realize the goal of assembly line, a standardized way of controlling target gene expression is necessary. Here we mainly concern the application of riboregulators, which was successfully introduced by [[http://www.openwetware.org/wiki/IGEM:UC_Berkeley/2006 Berkeley iGEM Team in year 2006]]. |
{|border=''1'' cellspacing=''1'' | {|border=''1'' cellspacing=''1'' | ||
|+ '''Original data''' | |+ '''Original data''' | ||
|- | |- | ||
| - | | Lock-3 | + | | Lock-3 (<bbpart>BBa_I714080</bbpart>) || 1.464e4 |
|- | |- | ||
| - | | Lock-1 | + | | Lock-1 (<bbpart>BBa_I714081</bbpart>) || 1.529e4 |
|- | |- | ||
| - | | GFP | + | | GFP (<bbpart>BBa_I7100</bbpart>) || 2.433e5 |
|- | |- | ||
| - | | | + | | Key-1 (<bbpart>BBa_I7140377</bbpart>) + Lock-1 || 1.198e5 |
|- | |- | ||
| - | | | + | | F-oriT- Key-1 (<bbpart>BBa_I714040</bbpart>) + Lock-1 || 1.697e4 |
|- | |- | ||
| - | | | + | | Key-2 (<bbpart>BBa_I714074</bbpart>) + Lock-1 || 4.663e4 |
|} | |} | ||
| Line 136: | Line 139: | ||
| Riboregulator pair || Open ratio | | Riboregulator pair || Open ratio | ||
|- | |- | ||
| - | | Lock-1 + Key-1 | + | | Lock-1 + Key-1 || 43% |
|- | |- | ||
| - | | Lock-1 + F-oriT- Key-1 | + | | Lock-1 + F-oriT- Key-1 || 0.7% |
|- | |- | ||
| - | | Lock-1 + Key-2 | + | | Lock-1 + Key-2 || 13% |
|} | |} | ||
| - | Conclusion: | + | |
| + | '''Conclusion:''' | ||
1. Attaching oriT to the key significantly reduces the Open ratio. | 1. Attaching oriT to the key significantly reduces the Open ratio. | ||
| Line 151: | Line 155: | ||
3. Relative open ratio is essentially the same despite the cell growth (data not shown). | 3. Relative open ratio is essentially the same despite the cell growth (data not shown). | ||
| + | ---- | ||
===Summary: How far can we push tandem conjugation=== | ===Summary: How far can we push tandem conjugation=== | ||
| - | We have constructed most mutant strains needed for tandem conjugation, as well as the signaling plasmids needed. Besides, we have measured the conjugation efficiency of various donor-recipient pairs, and tested the possibility of riboregulators as interface with other devices. All these preparation strongly suggest the feasibility of our hop count project. | + | *We have constructed most mutant strains needed for tandem conjugation, as well as the signaling plasmids needed. Besides, we have measured the conjugation efficiency of various donor-recipient pairs, and tested the possibility of riboregulators as interface with other devices. All these preparation strongly suggest the feasibility of our hop count project. |
| - | The final step towards our grail should be the construction of one unified strain containing multiple conjugative plasmids (all with relaxase deleted). Here it is relatively difficult to extract the mutated plasmids from constructed mutants and transform them into one strain, because of the large size of conjugative plasmid (50 – 100 kbp). We propose to transfer all conjugative plasmids into one recipient cell before deleting the oriT region and relaxase. Proper selective markers will be added to conjugative plasmids before transferring. | + | *The final step towards our grail should be the construction of one unified strain containing multiple conjugative plasmids (all with relaxase deleted). Here it is relatively difficult to extract the mutated plasmids from constructed mutants and transform them into one strain, because of the large size of conjugative plasmid (50 – 100 kbp). We propose to transfer all conjugative plasmids into one recipient cell before deleting the oriT region and relaxase. Proper selective markers will be added to conjugative plasmids before transferring. |
Latest revision as of 03:42, 27 October 2007
| Peking | The Projects--Hop-Count | The Team | Misc & Fun | [http://www.openwetware.org/wiki/IGEM:Peking/2007 Our OWW] | Acknowledgement |
|---|
Abstract
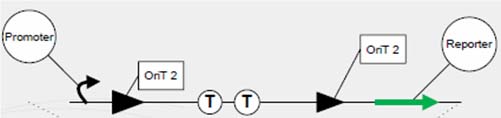
To meet the need of spatial differentiation in an assembly line, the hop count provides the number of network devices between the starting node and the destination node. We have developed the system of hop-count conjugation for cell-cell communication within a network of Escherichia coli. Here, the signals transferred are represented by DNA sequence, and record the times of transfer by losing tandem region at every conjugation event. To construct this system, tandem oriT pairs (BBa_J01002 and more) are inserted into signaling plasmid, and relaxase responsible for initiating conjugation events are removed from natural conjugation system and (attaching a terminator) inserted between these oriT pairs. At each single conjugation event, with the expression relaxase, the corresponding tandem oriT pair is lost with the rolling-cycle replication. Sequential conjugation and deletion of the different oriT pairs can be achieved by fine control of relaxase expression. The resulting tandem signaling conjugation process will allow us to develop networks of bacteria capable of sequentially assembling tasks.
Achievement
Measure conjugation efficiency of multiple conjugative strains
Construct conjugative mutant strains with oriT and relaxase deleted (and they WORK!)
Measure efficiency and specificity of multiple riboregulator pairs
![]() Construct signaling plasmid with tandem-oriT pairs
Construct signaling plasmid with tandem-oriT pairs
![]() Construct one unified strain containing multiple conjugation systems
Construct one unified strain containing multiple conjugation systems
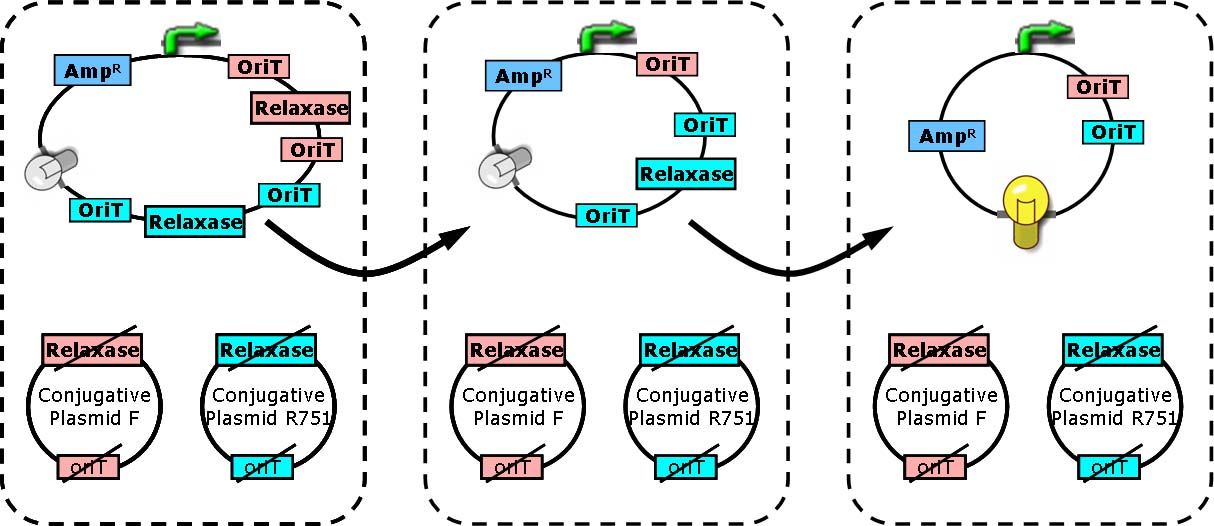
Design principle
Two key players of bacterial conjugation from donor cells to recipients we concern here are relaxase and oriT (origin of transfer). oriT is responsible for the initiation and termination of roll-cycling DNA replication during conjugation. The transfer of conjugative plasmid from one cell to another, in the form of single strand, starts from one half of oriT and ends at another half, separated by a “nick”. The nick site is generated by relaxosome, and relaxase is a major composition of it.
In cells with relaxase deleted, the process of conjugation is not supposed to happen due to the failure of nick site formation. Furthermore, if two oriT instead of one are located on the conjugative plasmid, the process of transferring DNA will start from one oriT and ends at another. Consequently, regions between two oriT without vegetative replication origin will be lost in recipient cells [Draper O et al., (2000). J Bacteriol 182:3191].
Each group of conjugative plasmid has specific relaxase and oriT. Therefore, the sequential conjugation process can be achieved by using one signaling plasmid with multiple sets of oriT pairs and relaxase (and terminators), and multiple conjugative plasmids with their relaxases deleted. All relaxases are controlled by the same promoter. As a result, the next relaxase expresses following the success of the former conjugation. To report the ending of tandem conjugation process, a reporter gene is attached after the last oriT pair, and hence expresses after all conjugation events happened.
Measure conjugation efficiency of strain with conjugative plasmid F
| Donor strain | Signaling plasmid | Conjugation efficiency |
| F-wild-type | one F-oriT | 0.050 |
| F-wild-type | a pair of F-oriT | 0.032 |
| F-oriT-Knockout | one F-oriT | 0.061 |
| F-oriT-Knockout | a pair of F-oriT | 0.037 |
- Conjugation efficiency: percentage of successful conjugation events to donor cells
Conclusion:
1. The deletion of oriT in wild type conjugative plasmid increases the conjugation efficiency of signaling plasmid by 20%.
2. A pair of oriT decreases the conjugation efficiency by 40%.
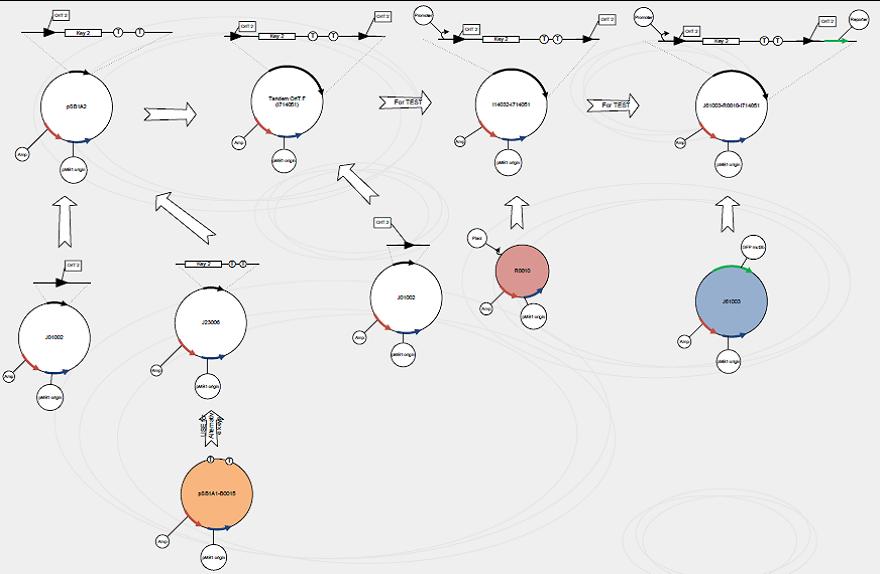
Construct signaling plasmid with tandem-oriT pairs
We use standard assembly protocols to construct signaling plasmid. To make sure every achievement is a solid progress to our goal, we designed a product map following industrial principle.
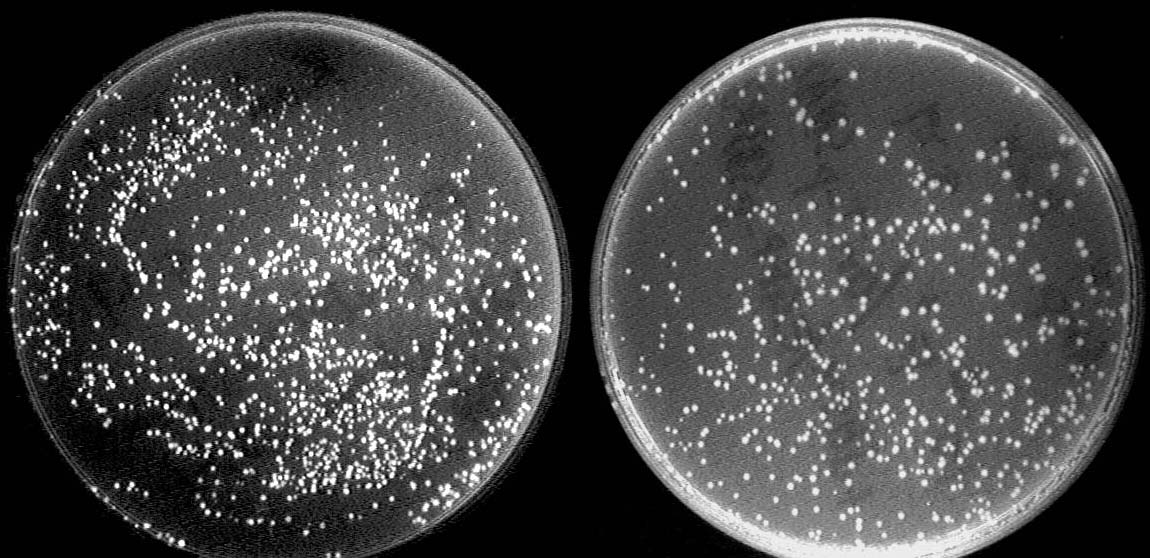
One outstanding achievement is that we have constructed BBa_I714061 and tested it with conjugation experiment, getting a positive result with the reporter gene expressed.
Left: After conjugation with a pair of oriT. The reporter GFP expresses due to the deletion of terminator.
Right: Control strain with no GFP expressed.
Conclusion:
1. Genes express properly in a signaling plasmid with the oriT pair deleted.
2. We successfully implemented and verified conjugation-deletion events with F system (binary hop count).
Construct conjugative mutant strains with oriT and relaxase deleted
Method 1: pKO3 system
1. Use plasmid pKO3 to generate precise deletions of oriT region in conjugative plasmids through homologous recombination.
2. For working principle of pKO3 please refer to: Link A. et al., (1997) J Bacteriol 179: 6228.
3. For more experimental details please visit [http://www.openwetware.org/wiki/IGEM:Peking/2007/Count:Knockout Knockout protocol in our OWW site].
Method 2: Lambda Red system
1. Use plasmid pKD46 and homologous PCR products to inactivate genes of relaxase in conjugative plasmids.
2. For working principle of Lambda Red system please refer to: Datsenko KA et al., (2000). Proc Natl Acad Sci USA 97: 6640
3. All experimental details are essentially the same as it is mentioned in the paper above.
Method 3: Reversed cyclic PCR
1. Used for in vitro construction of plasmid pSC101, a conjugative plasmid in size of 9 kbp.
2. For deletion a region in the plasmid, a pair of primers adjacent to the target region is first design (with the same restriction site), then mixed with plasmid pSC101 for PCR. The resulting product is self-ligated using single enzyme digestion condition and transferred into DH5a.
Achievement:
1. Successfully deleted genes of relaxase in plasmid F(BBa_I714035) , R751(BBa_I714037) and pSC101(BBa_I714038) .
2. BBa_I714035 successfully inactivated the conjugation process. BBa_I714037 has a leaky of about 1/10.
Measure efficiency and specificity of multiple riboregulator pairs
In order to provide an interface for interaction between our conjugation system and general genetic devices to realize the goal of assembly line, a standardized way of controlling target gene expression is necessary. Here we mainly concern the application of riboregulators, which was successfully introduced by http://www.openwetware.org/wiki/IGEM:UC_Berkeley/2006 Berkeley iGEM Team in year 2006.
| Lock-3 (BBa_I714080) | 1.464e4 |
| Lock-1 (BBa_I714081) | 1.529e4 |
| GFP (BBa_I7100) | 2.433e5 |
| Key-1 (BBa_I7140377) + Lock-1 | 1.198e5 |
| F-oriT- Key-1 (BBa_I714040) + Lock-1 | 1.697e4 |
| Key-2 (BBa_I714074) + Lock-1 | 4.663e4 |
| Riboregulator pair | Open ratio |
| Lock-1 + Key-1 | 43% |
| Lock-1 + F-oriT- Key-1 | 0.7% |
| Lock-1 + Key-2 | 13% |
Conclusion:
1. Attaching oriT to the key significantly reduces the Open ratio.
2. Unspecific Open ratio is relatively high, meaning a strong interaction among different riboregulators.
3. Relative open ratio is essentially the same despite the cell growth (data not shown).
Summary: How far can we push tandem conjugation
- We have constructed most mutant strains needed for tandem conjugation, as well as the signaling plasmids needed. Besides, we have measured the conjugation efficiency of various donor-recipient pairs, and tested the possibility of riboregulators as interface with other devices. All these preparation strongly suggest the feasibility of our hop count project.
- The final step towards our grail should be the construction of one unified strain containing multiple conjugative plasmids (all with relaxase deleted). Here it is relatively difficult to extract the mutated plasmids from constructed mutants and transform them into one strain, because of the large size of conjugative plasmid (50 – 100 kbp). We propose to transfer all conjugative plasmids into one recipient cell before deleting the oriT region and relaxase. Proper selective markers will be added to conjugative plasmids before transferring.