Caltech/Project/Recombineering
From 2007.igem.org
(→Background) |
|||
| (4 intermediate revisions not shown) | |||
| Line 18: | Line 18: | ||
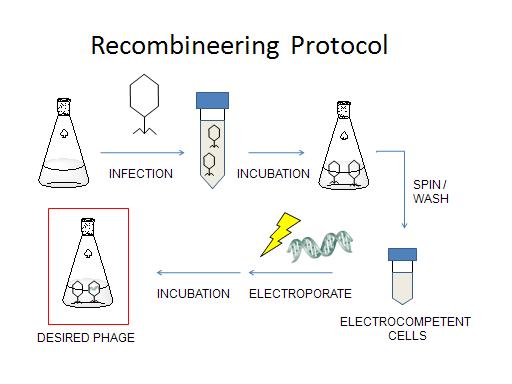
One can adapt recombineering to introduce short mutations into lambda phage genomes[1]. Briefly, ''E. coli'' cells expressing the proper prophage gene suite are infected with the lambda phage strain to be engineered. The cells are then made electrocompetent and single stranded oligos (about 70 nt) containing the desired mutation, plus flanking homology regions, are electroporated in. The new phages, a mixed population of wild type and engineered, are then allowed to lyse the host cells. The resulting lysate is screened in an appropriate manner to detect the mutation of interest. | One can adapt recombineering to introduce short mutations into lambda phage genomes[1]. Briefly, ''E. coli'' cells expressing the proper prophage gene suite are infected with the lambda phage strain to be engineered. The cells are then made electrocompetent and single stranded oligos (about 70 nt) containing the desired mutation, plus flanking homology regions, are electroporated in. The new phages, a mixed population of wild type and engineered, are then allowed to lyse the host cells. The resulting lysate is screened in an appropriate manner to detect the mutation of interest. | ||
| - | [[Image: | + | [[Image:Recombineering_Protocol1.jpg|center]] |
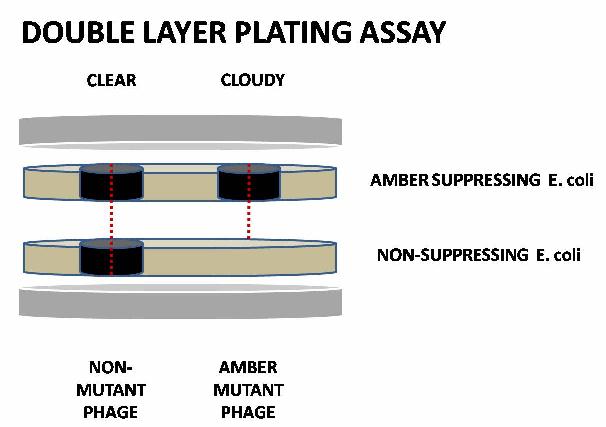
In our particular application, we are introducing amber stop codons in frame to <i>N</i> and <i>Q</i>. To screen for these mutations, phages are plated for plaques in an amber suppressor strain (E. coli LE392, <i>supF</i>). A further layer of non-amber suppressing cells (E. coli MG1655), without phages, is immediately plated on top of the amber suppressing layer. Phages without the amber mutation will make plaques that 'punch through' both layers, while amber mutant phages will be unable to penetrate the upper layer. The two types of plaques can be distinguished by eye, allowing us to screen for amber mutants. | In our particular application, we are introducing amber stop codons in frame to <i>N</i> and <i>Q</i>. To screen for these mutations, phages are plated for plaques in an amber suppressor strain (E. coli LE392, <i>supF</i>). A further layer of non-amber suppressing cells (E. coli MG1655), without phages, is immediately plated on top of the amber suppressing layer. Phages without the amber mutation will make plaques that 'punch through' both layers, while amber mutant phages will be unable to penetrate the upper layer. The two types of plaques can be distinguished by eye, allowing us to screen for amber mutants. | ||
| Line 40: | Line 40: | ||
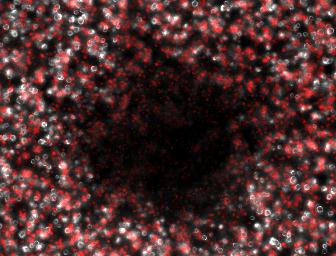
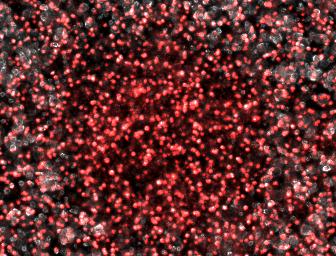
We therefore attempted a slight modification, labeling the non-suppressing bacterial with a high copy plasmid driving an RFP. This process did indeed make the two plaque types more distinguishable with the aid of a fluorescence dissecting scope. Microscopy results confirm that the two plaque types do indeed correspond to different degrees of lysis in the two layers. Below on the left is an obligate lytic mutant (cI60), which creates plaques in both unlabeled suppressor (white) and RFP labeled non-suppressor cells (red). Below on the right is an N-amber mutant phage creating a plaque in the unlabeled suppressor cells, while leaving the RFP labeled non-suppressor cells intact. | We therefore attempted a slight modification, labeling the non-suppressing bacterial with a high copy plasmid driving an RFP. This process did indeed make the two plaque types more distinguishable with the aid of a fluorescence dissecting scope. Microscopy results confirm that the two plaque types do indeed correspond to different degrees of lysis in the two layers. Below on the left is an obligate lytic mutant (cI60), which creates plaques in both unlabeled suppressor (white) and RFP labeled non-suppressor cells (red). Below on the right is an N-amber mutant phage creating a plaque in the unlabeled suppressor cells, while leaving the RFP labeled non-suppressor cells intact. | ||
| - | [[Image:DoubleLayerCompositecI60.jpg| | + | [[Image:DoubleLayerCompositecI60.jpg|250px|left|frame|<center>Clear Plaque</center>]] |
| + | [[Image:DoubleLayerCompositeNam.jpg|none|250px|frame|<center>Cloudy Plaque</center>]] | ||
| + | |||
==Relevant Protocols== | ==Relevant Protocols== | ||
Latest revision as of 05:54, 27 October 2007
iGEM 2007
Home
Highlights
Project
People
Protocols
|