Caltech/Project/Q
From 2007.igem.org
| (One intermediate revision not shown) | |||
| Line 15: | Line 15: | ||
==Role in Lambda Life Cycle== | ==Role in Lambda Life Cycle== | ||
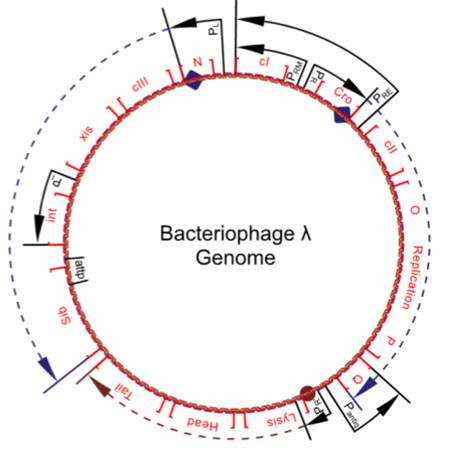
| + | [[Image:Bacteriophage_lambda_genome.jpg|right|thumb|Schematic representation of the lambda genome. Q alters RNA polymerase and allows transcription of genes coding for heads and tails production.]] | ||
Since Q is located downstream of the terminator tR, its transcription requires the presence of N. During early stages of lambda's life cycle, as well as in the lysogenic pathway, the translation of Q is downregulated by an antisense RNA. The promoter pAQ transcribes Q in reverse, producing a Q-antisense RNA that binds to the Q messenger RNA and prevents ribosomal attachment. Furthermore, a relatively high concentration of anti-Q is required to effectively cause antitermination, giving the phage time to begin a lysogenic life cycle if conditions are appropriate. | Since Q is located downstream of the terminator tR, its transcription requires the presence of N. During early stages of lambda's life cycle, as well as in the lysogenic pathway, the translation of Q is downregulated by an antisense RNA. The promoter pAQ transcribes Q in reverse, producing a Q-antisense RNA that binds to the Q messenger RNA and prevents ribosomal attachment. Furthermore, a relatively high concentration of anti-Q is required to effectively cause antitermination, giving the phage time to begin a lysogenic life cycle if conditions are appropriate. | ||
Once Q has modified the RNA polymerase, the polymerase begins to transcribe genes beyond tR'. These genes are crucial to the production of new phage components, such as tails and heads (protein capsules). Once these genes are expressed, the phage is committed to the lytic pathway. | Once Q has modified the RNA polymerase, the polymerase begins to transcribe genes beyond tR'. These genes are crucial to the production of new phage components, such as tails and heads (protein capsules). Once these genes are expressed, the phage is committed to the lytic pathway. | ||
| - | |||
| - | |||
==Original Goals== | ==Original Goals== | ||
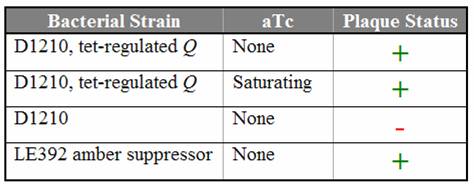
| - | The goal of this subproject is to determine the approximate concentration of Q necessary to | + | The goal of this subproject is to determine the approximate concentration of Q necessary to allow infection. The Q gene cloned into a tetracycline-inducible expression system as described [[Caltech/Project/Constructs|here]]. ''E. coli'' cells containing the Q construct were titered with ''Q'' amber mutants, and the production of Q in the host cell was controlled by varying the concentration of the inducer aTc. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ==Results== | |
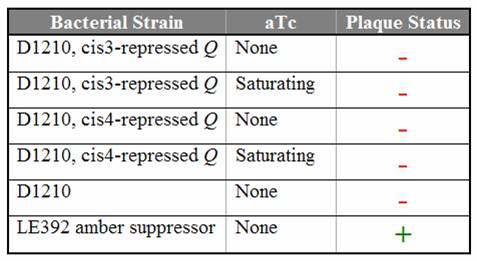
| + | Titration trials with Q amber mutants have shown that expression of Q under tetR repression is leaky. In the absence of aTc, we still observed lysis using Q amber mutant phage. Replacing the spacer with various [[Caltech/Project/Riboregulator|cis-repressor]] sequences has shut down the leaky expression. Even in the presence of saturating aTc, no lysis was observed using the cis3 and cis4 repressed Q constructs. | ||
| - | [[Image: | + | [[Image:Q_results.jpg|center|frame|Titering results using the Q amber mutant on D1210 ''E. coli'' containing the Q construct]] |
| + | [[Image:cisQ_results.jpg|center|frame|Titering results using the Q amber mutant on D1210 ''E. coli'' containing the cis-repressed Q constructs]] | ||
| + | We are currently working to introduce the conjugate trans activators to the cis3-repressed Q construct. | ||
|} | |} | ||
</div> | </div> | ||
Latest revision as of 00:46, 27 October 2007
iGEM 2007
Home
Highlights
Project
People
Protocols
|