Duke/Projects/bp
From 2007.igem.org
EricJosephs (Talk | contribs) |
EricJosephs (Talk | contribs) |
||
| (3 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
==Parts and setup== | ==Parts and setup== | ||
In order to optimize the experimental gene circuit and better control the ratio of 4-hydroxybutyrate (largely represented in the copolymer) to 3-hydroxybutyrate, we modeled the system in order to locate the limiting | In order to optimize the experimental gene circuit and better control the ratio of 4-hydroxybutyrate (largely represented in the copolymer) to 3-hydroxybutyrate, we modeled the system in order to locate the limiting | ||
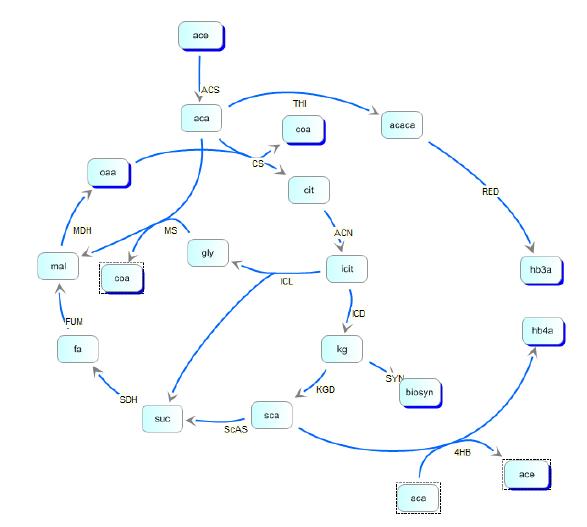
| - | reaction steps in the metabolic pathway of E. coli and in the polymer producing extension of those pathways. | + | reaction steps in the metabolic pathway of E. coli and in the polymer producing extension of those pathways. We then created the model depicted below: |
| - | + | ||
| - | + | [[Image:bp1.jpg]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Kinetic pathways for 3HB and 4HB were developed and analyzed using integrated tools | Kinetic pathways for 3HB and 4HB were developed and analyzed using integrated tools | ||
from the Systems Biology Workbench (SBW). Jdesigner2 was used to build the network | from the Systems Biology Workbench (SBW). Jdesigner2 was used to build the network | ||
| Line 19: | Line 16: | ||
Iadevaia, et. al. Various other miscellaneous enzyme rates were obtained from publicly | Iadevaia, et. al. Various other miscellaneous enzyme rates were obtained from publicly | ||
available databases such as BRENDA. | available databases such as BRENDA. | ||
| - | |||
| - | |||
==Results== | ==Results== | ||
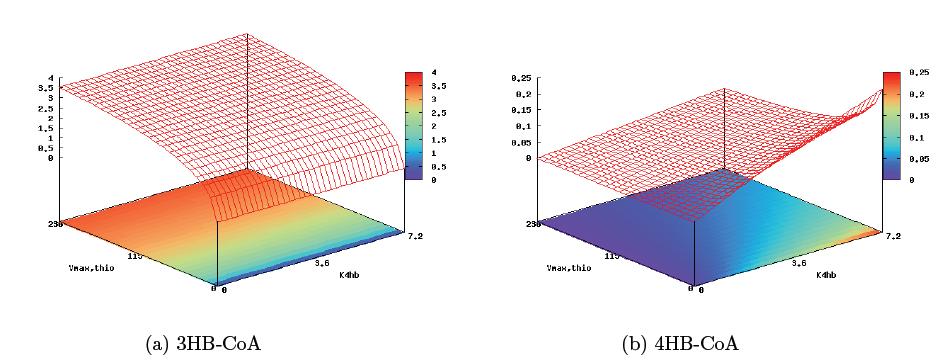
| - | + | With these derivations, it is now possible to analyze the pathway and predict key control | |
| + | sites that will allow for copolymer regulation. The pathway structure seems to suggest the | ||
| + | 4HB reaction step and the first step of the 3HB reaction as key enzymatic steps harboring | ||
| + | the greatest effect on copolymer composition. Plotting reaction rates of monomer conversion | ||
| + | against the maximum enzyme rate in the first step of 3HB formation, Vmax,thio (0–229mmol | ||
| + | min−1) and the 4HB parameter K4HB (0–7.2mmol min−1), gives a sense of the level of control | ||
| + | two independent inducible promoters would provide us in this circuit (the figure below). From the | ||
| + | graph, it is immediately apparent that regulation of the 4HB enzymatic rate is only able | ||
| + | to control the rate of 4HB monomer formation, while regulation of the 3HB enzmatic rate affects both monomer formation dynamics. This model then suggests that in building a poly(3HB-co-4HB) pathway, the the best regulation target would be the thiolase enzyme of the 3HB monomer pathway, and the level of pathway induction would be medium strength, allowing for significant 4HB accumulation while still keeping overall polymer yield high. | ||
| + | |||
[[Image:bp2.jpg]] | [[Image:bp2.jpg]] | ||
| - | |||
Latest revision as of 03:31, 27 October 2007
Contents |
Bioplastics Synthesis
Background and Motivation
With rising concerns over environmental pollution and energy shortages, there has been a greater push towards alternative energy and environmentally friendly production of useful materials. Currently, the production of petrochemical plastics not only ejects hazardous gases into the atmosphere but also generates considerable amounts of waste. Along with a strong desire to become independent of oil-based products, these concerns have paved the way for research into highly biodegradable and biocompatible bioplastics, such as polyhydroxyalkanoates (PHAs), a particular polymer that is formed natively in several strains of bacteria. This project seeks to improve the production of a certain PHA, poly(3-hydroxybutyrate-co-4-hydroxybutyrate, through the maximization of the desirable 4-hydroxybutyrate monomer. This is because a high ratio of 3-hydroxybutyrate produces a very crystalline and brittle plastic while high 4-hydroxybutyrate produces a very desirable elastomers.
Parts and setup
In order to optimize the experimental gene circuit and better control the ratio of 4-hydroxybutyrate (largely represented in the copolymer) to 3-hydroxybutyrate, we modeled the system in order to locate the limiting reaction steps in the metabolic pathway of E. coli and in the polymer producing extension of those pathways. We then created the model depicted below:
Kinetic pathways for 3HB and 4HB were developed and analyzed using integrated tools from the Systems Biology Workbench (SBW). Jdesigner2 was used to build the network and deterministic modeling and steady-state analysis were performed using Jarnac[24]. Reaction kinetics of the tricarboxyilic acid cycle were reconstructed from the work of Singh, et. al., while reaction rates and kinetics for 3HB kinetics were obtained from the work of Iadevaia, et. al. Various other miscellaneous enzyme rates were obtained from publicly available databases such as BRENDA.
Results
With these derivations, it is now possible to analyze the pathway and predict key control sites that will allow for copolymer regulation. The pathway structure seems to suggest the 4HB reaction step and the first step of the 3HB reaction as key enzymatic steps harboring the greatest effect on copolymer composition. Plotting reaction rates of monomer conversion against the maximum enzyme rate in the first step of 3HB formation, Vmax,thio (0–229mmol min−1) and the 4HB parameter K4HB (0–7.2mmol min−1), gives a sense of the level of control two independent inducible promoters would provide us in this circuit (the figure below). From the graph, it is immediately apparent that regulation of the 4HB enzymatic rate is only able to control the rate of 4HB monomer formation, while regulation of the 3HB enzmatic rate affects both monomer formation dynamics. This model then suggests that in building a poly(3HB-co-4HB) pathway, the the best regulation target would be the thiolase enzyme of the 3HB monomer pathway, and the level of pathway induction would be medium strength, allowing for significant 4HB accumulation while still keeping overall polymer yield high.