ETHZ
From 2007.igem.org
(→Lab Work) |
(→Lab Schedule) |
||
| Line 121: | Line 121: | ||
=== Lab Schedule === | === Lab Schedule === | ||
<font face="DejaVu Sans" size="3"><b>Week 1</b></font> <b>06.08.07 - 12.08.07</b><br> | <font face="DejaVu Sans" size="3"><b>Week 1</b></font> <b>06.08.07 - 12.08.07</b><br> | ||
| - | [[ETHZ/labnotes| | + | [[ETHZ/labnotes| lab schedule week 1]]<BR> |
<font face="DejaVu Sans" size="3"><b>Week 2</b></font> <b>13.08.07 - 19.08.07</b><br> | <font face="DejaVu Sans" size="3"><b>Week 2</b></font> <b>13.08.07 - 19.08.07</b><br> | ||
| - | [[ETHZ/labnotes| | + | [[ETHZ/labnotes| lab schedule week 2]]<BR> |
<font face="DejaVu Sans" size="3"><b>Week 3</b></font> <b>20.08.07 - 26.08.07</b><br> | <font face="DejaVu Sans" size="3"><b>Week 3</b></font> <b>20.08.07 - 26.08.07</b><br> | ||
| - | [[ETHZ/labnotes| | + | [[ETHZ/labnotes| lab schedule week 3]]<BR> |
<font face="DejaVu Sans" size="3"><b>Week 4</b></font> <b>27.08.07 - 02.09.07</b><br> | <font face="DejaVu Sans" size="3"><b>Week 4</b></font> <b>27.08.07 - 02.09.07</b><br> | ||
| - | [[ETHZ/labnotes| | + | [[ETHZ/labnotes| lab schedule week 4]]<BR> |
=== Lab Notes / Log Book === | === Lab Notes / Log Book === | ||
Revision as of 19:57, 13 August 2007

Contents |
Team Members
| [[Image:|thumb|398px|Picture: The ETH Zuerich iGEM Team 2007 (left to right: )]] |
For the International Competition on Genetically Engineered Machines, the teams should be composed of both biologist and engineers. That way, the engineers will try to develop new systems in a bottom-up fashion and run numerical simulations, while the biologists will be able to assess the feasibility of such systems, and construct them from biological parts.
Instructors:
Joerg Stelling
Students:
Martin Brutsche
Katerina Dikaiou
Graduate Students:
[http://www.tik.ee.ethz.ch/~sop/people/thohm/ Tim Hohm]
Christian Kemmer
Introduction to Synthetic Biology
In order to get an initial understanding of the concepts of synthetic biology, we read and presented publications on various topics. A representative list of the topics that we covered on this “boot-camp” is listed in the following:
- Introduction to synthetic biology
- DNA de novo design
- DNA circuits
- Hysteresis
- Oscillators
- Zinc fingers
- Noise in single cell measurements
- Distance communication
- Parameter manipulations
- Protein logic
- Orthogonal systems
- mRNA engineering
- RNA regulators
Choosing the Project
Step 1: Brainstorming
Initially, we wanted to come up with as many ideas as possible, in order to be able to choose among them the best, and find a cool project to carry out. For this reason, we had brainstorming sessions. We split up in three teams, and each team tried to come up with many fancy and showy ideas, which was facilitated by keeping in mind the following brainstorming guidelines:
- Defer judgment - the rules of nature don't apply
- Encourage wild ideas
- Build on the ideas of others
- Be visual
- Go for quantity
- Stay focused on topic
The ideas that we came up with, as were presented in the following meetings, were:
- PID Controller: Design a PID controller out of biological elements. The P component can be a simple output to a regulatory protein, and the I component can be the overall protein production at a time period. What can the D component be?
- Motion Detector: Cells are grown on a petri dish. Below the dish, moving images are displayed. A 3-state automaton is proposed. Output A is created when light is present. Output B is created when light is absent. Moving patterns will cause some cells to create both outputs over time. This will result in some “inspector” cells producing output C, by collecting outputs A and B.
- Analog-to-Digital Converter: Compare the level of protein concentration with thresholds, and digitize the output.
- Neural Network: Create a sort of biological neural network with bacteria. We should address the issue of learning, and find a way to incorporate the feedback in the cell decision making process. Directed evolution can be a sort of feedback, but we want to avoid this. (This idea was the basis for the “learning project”)
- Paramedic Cells: Some cells are able to detect signals coming from other cells, and create food for them, or create proteins in order to save them and make them function better.
- Cell Batteries: Cells are able to create and store large quantities of ATP, during a “storing process”. Afterwards, they can detect a signal and give back all the energy they stored, in a short burst, like a capacitor. Other ideas are that the cells can “blow up” and emit large amounts of GFP, based on the ATP that they have accumulated.
- Flashing Bacteria: Cells are grown on a light pattern. The cells that are on the bright parts of the image are oscillating in phase, while the others are remaining dark. This results in the observation of a flashing pattern.
- Biocam: Visible to Fluorescent light converter.
- BioCD: “Print” cells on a film, then read them out and “reconstruct” the original data. Basically, it is an analog to digital converter, followed by a system that can interpret the digitized data. (This idea was the basis for the “Music of life project”, where cells would produce fluorescent proteins based on an analog input. Then, the amount and type of fluorescence would code some music).
- Clock: A follow-the-leader system. We have to groups of cells. The first group creates something that repels the second group. The second group creates a protein that attracts the first group. This way, they first group wants to “catch” the second group, whereas the second group wants to “avoid” the first group. This results in them moving around. We can say that the second group is the leader, and the first group exhibits a "follow-the-leader” behavior.
- Sensors: Various systems that can sense PH, pressure, temperature, meat quality, moisture e.t.c. have been proposed.
Step 2: Preferred Projects
We decided that three of the above ideas were worth a deeper examination. Namely, we split again in three teams, so that we can come up with an initial system, with remarks on its feasibility and coolness. Our results were presented on all the team members, so that we could then limit the potential projects down to two and one, and proceed with constructing the DNA sequence. The three projects and the presentations that we did were:
- Music of Life: The basic idea is that instead of having an analog-to-digital converter with four outputs (three fluorescent proteins, and no output), we can have two switches. When switch A is on, RFP is produced. When switch B is on, GFP is produced. When both switches A and B are on, a yellowish output is observed. By recording these outputs, we can later create music, by assigning each fluorescent protein to a chord. For example, RFP would correspond to a G chord. The strength of the fluorescence can signify the strength of the chord. If the cells are placed on a spinning disk, we can have something like a vinyl player. A camera is observing the cells, and music is created on the fly.
- Learning: Based on the idea of the neural network, we want to create a biological system, where the cells can learn a specific behavior. In order to simplify the system, we decided that the cells can learn to recognize a specific type of other cells. We divide the process in a learning phase, and a recognition phase. First, cells A are put together with cells B. Then, cells A are “learning” to recognize cells B. If afterwards they are put in a petri dish with cells B, they will emit GPF. Otherwise, they will stay dark.
Idea two, even though it was fancy, was fairly simple, as, creating two switches is a straightforward and well understood process. As a result, it was discarded. We could try to make the system more complex, but making something more complex just for the shake of it, is not a good engineering approach ;)
The Project
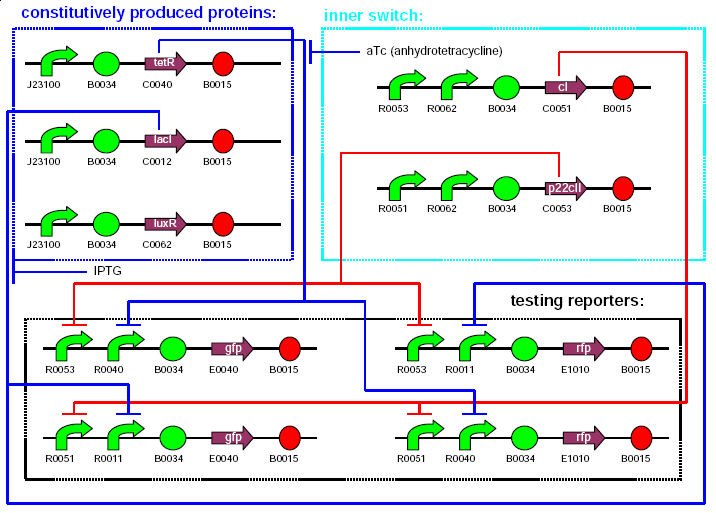
We decided to proceed with the Learning Project. Our goal is to design an E.coli strain, able to distinguish between two chemical substances after an assigned learning process, induced by an external chemical signal (teaching substance). After the bacterial strain was taken to a testing phase, the output will result in either yellow or cyan fluorescence, depending on whether the bacteria recognized the same chemical substance in the testing phase as it was trained in the learning phase or a different one. We aim to implement our system using a toggle switch consisting of different repressor and activator proteins coupled with double regulatory regions.
System Models
Simulations
detailed information on the simulations
Sensitivity Analysis
Lab Work
Here we can write when each one will be available. For example:
- Christos:
Can be available everyday after 17:30, till 00:00. Weekends as well. Also 11:00-13:00 everyday. Away 17.08-19.08, 22.08-24.08. - Tim:
13.08.-15.08. after 17:30
away from 17.08. until 02.09. - Martin:
Every day, also weekends, for half a day or even more if it's requiered. The last week of August and the first one in September I have only time for aproximately 5-6 h a day due to learning for the exams.
Cloning Plan
Lab Instructions and Methods
Lab Schedule
Week 1 06.08.07 - 12.08.07
lab schedule week 1
Week 2 13.08.07 - 19.08.07
lab schedule week 2
Week 3 20.08.07 - 26.08.07
lab schedule week 3
Week 4 27.08.07 - 02.09.07
lab schedule week 4
Lab Notes / Log Book
Here we will write down every day a detailed version of the work we have done (it should be the same stuff we write in the "real lab book" which stays in the lab). In this way everyone can see in detail what was done in the lab.