Edinburgh/DivisionPopper/Applications
From 2007.igem.org
Luca.Gerosa (Talk | contribs) |
|||
| Line 3: | Line 3: | ||
[[Edinburgh]] > '''DivisionPopper''' | [[Edinburgh]] > '''DivisionPopper''' | ||
| - | [[Edinburgh/DivisionPopper| Introduction]] | [[Edinburgh/DivisionPopper/Applications|Applications]] | [[Edinburgh/DivisionPopper/Design|Design]] | [[Edinburgh/DivisionPopper/Status|Status]] | [[Edinburgh/DivisionPopper/References|References]] | + | '''MENU''' :[[Edinburgh/DivisionPopper| Introduction]] | [[Edinburgh/DivisionPopper/Applications|Applications]] | [[Edinburgh/DivisionPopper/Design|Design]] | [[Edinburgh/DivisionPopper/Modelling|Modelling]] | [[Edinburgh/DivisionPopper/Status|Status]] | [[Edinburgh/DivisionPopper/References|References]] |
=Applications and alternate versions of division analysers= | =Applications and alternate versions of division analysers= | ||
Revision as of 12:15, 13 September 2007
https://static.igem.org/mediawiki/2007/f/f5/800px-Edinburgh_City_15_mod.JPG
Edinburgh > DivisionPopper
MENU : Introduction | Applications | Design | Modelling | Status | References
Applications and alternate versions of division analysers
Contents |
This page details some potential uses for the Division PoPper and other division analysis devices.
Division Frequency Analysis
The output of the Division PoPper could be linked to the production of a slowly degrading protein. The more frequent the divisions, the greater the concentration of the protein.
Division Counting
Coupling to a PoPS counting device
Couple the output of the Division PoPper to another counting device (such as the ETH Zurich counter or other variants) to count the number of cell divisions. This is difficult to test due to the nature of colonies and cells dividing out of phase. We get around this problem by using high-power microscopy to study the activity of single cells.
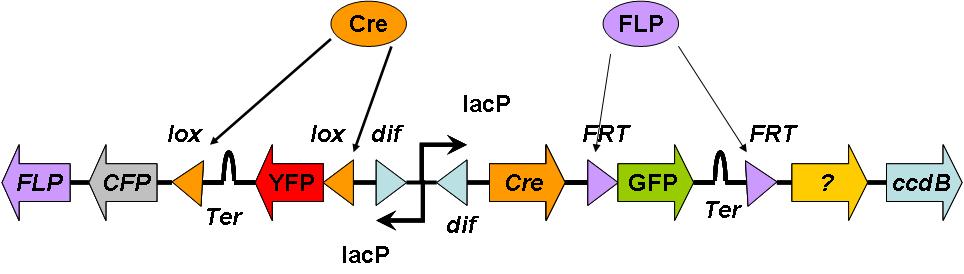
Counting using more recombination
Rather than using the DivisionPoPper directly, this uses flipping dif sites to activate different recombinases, cut out sections of DNA and thus enable a range of downstream functions with each division. Functions are represented by fluorescent reporter genes here for sake of visibility, but may be replaced with genes to execute a function of choice; e g metabolite receptors and directed cellular movement.
Counting at the mercy of lac operators
An divison-induced oscillator is constructed using genes with various numbers of lac operators upstream of them. LacI production is turned off and each cell division divides the remaining LacI protein amongst daughter cells. Thus gene functions are orderly induced as a function of the amount of upstream lac operators. Finally LacI production is induced and the process repeats.
Introduction | Applications | Design | Status | References