USTC/Logic-Gate Promoters
From 2007.igem.org
m |
m |
||
| Line 7: | Line 7: | ||
# They will never present in complicated structures; | # They will never present in complicated structures; | ||
# They will never include the restriction enzyme cutting sites that will be involved in the whole study; | # They will never include the restriction enzyme cutting sites that will be involved in the whole study; | ||
| - | # | + | # They will never include the recognition sites of RNA Polymerases and those of either of the two repressors. |
We plan to try N different positions for Operator A and M ones for Operator B, thus constructing N*M kinds of promoters which actually constitute a specific promoter family. After transformed into the four test plasmids (pCR00, pCR01, pCR10, pCR11) together with a downstream double-reporter system, we will get to know the solo-repression and co-repression effects of the two repressors on the specific promoter. | We plan to try N different positions for Operator A and M ones for Operator B, thus constructing N*M kinds of promoters which actually constitute a specific promoter family. After transformed into the four test plasmids (pCR00, pCR01, pCR10, pCR11) together with a downstream double-reporter system, we will get to know the solo-repression and co-repression effects of the two repressors on the specific promoter. | ||
Revision as of 14:41, 4 October 2007
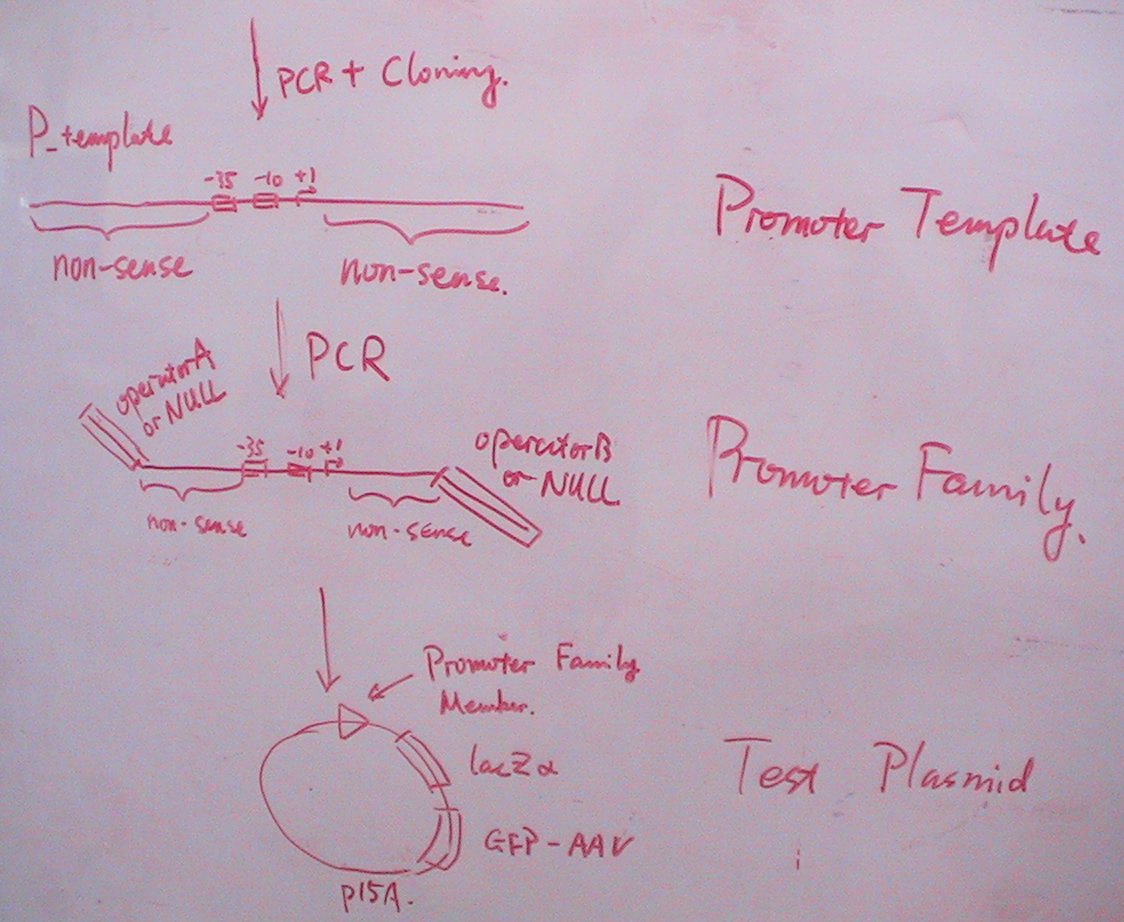
By means of PCR and molecular cloning, we come to possess the promoters that will serve as templates for the promoter family. There are several conservative sequences in the promoters, for instance, the -35 box, the -10 box, +1 start.
Moreover, there are two specific non-sense sequences in every promoter, one on each side of the transcription initiation site. The artificially added non-sense sequences are set once selected from random groups. They have three main characters:
- They will never present in complicated structures;
- They will never include the restriction enzyme cutting sites that will be involved in the whole study;
- They will never include the recognition sites of RNA Polymerases and those of either of the two repressors.
We plan to try N different positions for Operator A and M ones for Operator B, thus constructing N*M kinds of promoters which actually constitute a specific promoter family. After transformed into the four test plasmids (pCR00, pCR01, pCR10, pCR11) together with a downstream double-reporter system, we will get to know the solo-repression and co-repression effects of the two repressors on the specific promoter.