Davidson Missouri W
From 2007.igem.org
(→Western Meeting Notes) |
(→Western Meeting Notes) |
||
| Line 35: | Line 35: | ||
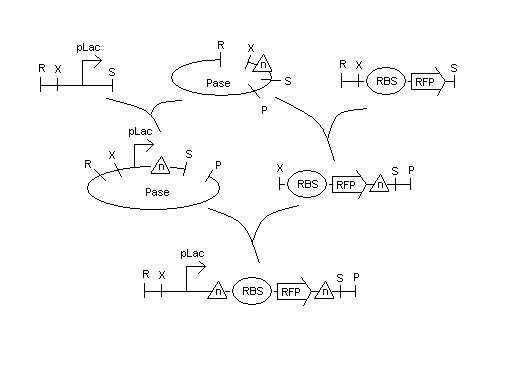
We have cultures that were grown overnight of the Hix(n). We received the sequencing data on the Karen's and Bruce's Hix(n) constructs (from 5/14 report) and are analyzing the data to find the mutations. Lab work continues. | We have cultures that were grown overnight of the Hix(n). We received the sequencing data on the Karen's and Bruce's Hix(n) constructs (from 5/14 report) and are analyzing the data to find the mutations. Lab work continues. | ||
| - | [[Image:hixn.jpg|thumb| | + | [[Image:hixn.jpg|thumb|600px|center| ]] |
''5/17'' | ''5/17'' | ||
Revision as of 16:00, 18 May 2007
===Davidson & Missouri Western Team Logos, iGEM2006===
Contents |
Western Meeting Notes
5/14 Overview of goals for the next two weeks
Begin Experimentation
Educational Information
Literature Research
5/15
Reviewed PowerPoint presentations of 2006 project to gain an understanding of the Pancake Problem.
Viewed Bruce's presentation on the mutation of the hix sites
Developed plan to build testing constructs of mutant hix sites, arriving this afternoon from Davidson by FedEx
Will build pLac - hix(n) - BBa_S03644 (RFPrev-RBSrev) - hix(n) on AmpR plasmid. Details to follow.
Note -- The above plan has changed as of Tuesday afternoon because of availability of parts.
BBa_S03644 is not available in our lab and the exact forward equivalent appears (amazingly) to not exist anywhere. However, there is an RBS - RFP in the Registry (BBa_I13502) which is a preassembled combination of a weaker RBS (BBa_B0034) and RFP (BBa_E1010). There is also a full positive control assembly (BBa_J04450) of pLac - B0034 - E1010 - Term. We will need to be careful to use the same parts in our Hix(n) assemblies so that we can test for only the functionality of our construct with the mutated Hix sites included.
5/16
We have cultures that were grown overnight of the Hix(n). We received the sequencing data on the Karen's and Bruce's Hix(n) constructs (from 5/14 report) and are analyzing the data to find the mutations. Lab work continues.
5/17
Karen developed a modified Green Fuorescent protein, GFP, by splitting the DNA for GFP, reversing one portion and adding Hix sites to it. This way, GFP can only be produced when the reversed portion is inverted so that the entire gene is in the correct order. This method was attempted with the TET resistance gene with no results. We have looked at using several fluorescent proteins modified in this manner to study the Traveling Salesman. In this manner, the proteins would only be expressed if they had all been lined up in the correct order. This approach has some flaws however, all fluorescent proteins are similar in structure, so that even if the portions of two different genes come together, the resulting protein may still fluoresce. Additionally, fluorescent protiens have the ability to undergo fluorescent resonant energy transfer, FRET, a phenomenon in which energy absorbed by one molecule is directly transfered to a second molecule, this prevents the first molecule from emitting visible photons, so that it may remain undetected.
Students
• Jordan Baumgardner, junior biology, biochem/molecular bio major, [1]
• Ryan Chilcoat, junior biology major (health sciences), [2]
• Tom Crowley, senior biochem/molecular biology major, [3]
• Lane H. Heard, Central High School graduate, [4]
• Nickolaus Morton, junior chemistry major, [5]
• Michelle Ritter, junior mathematics major, [6]
• Miranda Showalter, junior biology major (health sciences), [7]
• Jessica Treece, junior biology major (health sciences), [8]
• Matthew Unzicker, senior biochem/molecular biology major, [9]
• Amanda Valencia, senior biochem/molecular biology major, [10]
• Will DeLoache, ??? major, [11]
• Oyinade Adefuye, ??? major, [12]
• Jim Dickson , ??? major, [13]
• Amber Shoecraft, ??? major, [14]
• Andrew Martens, ??? major, [15]
• Michael Waters, ??? major, [16]
Faculty
• Todd Eckdahl [http://staff.missouriwestern.edu/~eckdahl/], Professor, Department of Biology, [17]
• Jeff Poet [http://staff.missouriwestern.edu/~poet/], Assistant Professor, Department of Computer Science, Mathematics, and Physics, [18]
Shipping Address: Todd Eckdahl, Biology Department, Missouri Western State University, 4525 Downs Drive, Saint Joseph, MO, 64507 [(816) 271-5873]
• Malcolm Campbell [http://www.bio.davidson.edu/people/macampbell/macampbell.html], Professor, Department of Biology, [19]
• Karmella Haynes [http://www.bio.davidson.edu/people/kahaynes/kahaynes.html], Visiting Assistant Professor, Department of Biology, [20]
• Laurie Heyer [http://www.davidson.edu/math/heyer/], Associate Professor, Department of Mathematics, [21]
Shipping Address: Malcolm Campbell, Biology Dept. Davidson College, 209 Ridge Road, Davidson, NC 28036 [(704) 894-2692]
Project Overview
Solving the Pancake Problem with an E. coli Computer Our goal is to genetically engineer a biological system that can compute solutions to a puzzle called the burnt pancake problem. The EHOP computer is a proof of concept for computing in vivo, with implications for future data storage devices and transgenic systems.
Resources / Citations
Cool site for Breakfast [http://www.cut-the-knot.org/SimpleGames/Flipper.shtml]
Karen Acker's paper describing GFP and TetA(c) with Hix insertions [http://www.bio.davidson.edu/Courses/Immunology/Students/spring2006/Acker/Acker_finalpaperGFP.doc]
Bruce Henschen's paper describing one-time flippable Hix sites [http://www.bio.davidson.edu/Courses/genomics/2006/henschen/Bruce_Finalpaper.doc]
Intro to Hamiltonian Path Problem and DNA [http://www.ams.org/featurecolumn/archive/dna-abc2.html]