NYMU Taipei/Lab Notes/2007 9 16
From 2007.igem.org
(Difference between revisions)
Lihsiangyen (Talk | contribs) (→Gel separation) |
Lihsiangyen (Talk | contribs) (→Enzyme Digestion) |
||
| Line 17: | Line 17: | ||
** H2O: 6ul | ** H2O: 6ul | ||
** Total: 20ul | ** Total: 20ul | ||
| + | *Enzyme digestion: D-term E with XbaI | ||
| + | **D-term E: 30ul | ||
| + | **XbaI: 2ul | ||
| + | **10x 2 buffer: 5ul | ||
| + | **10x BSA: 5ul | ||
| + | **H2O: 8ul | ||
| + | **Total: 50ul | ||
==Gel separation== | ==Gel separation== | ||
Revision as of 11:01, 16 September 2007
Enzyme Digestion
- Enzyme digestion: CinR+HSL (with RBS) with EcoRI, SpeI
- CinR+HSL (1~4): 14ul
- 因為濃度未知, 所以用最大體積
- EcoRI: 1ul (20,000 units/ml)
- SpeI: 1ul (10,000 units/ml)
- 10x EcoRI buffer: 2ul
- Total: 20ul
- CinR+HSL (1~4): 14ul
- Enzyme digestion: D-term with EcoRI
- Original we need double digestion EcoRI and XbaI.
- However, NEB recommend to perform the digestion in sequential manner.
- Thus, we digest the EcoR1 first
- D-term: 11ul (損失因子10, 目標重量300ng)
- 300(ng) * 10 = 3(ug) = 0.27 (ug/uL) * X(uL), X = 3/0.27 = 11.11111111
- EcoRI: 1ul
- 10x EcoRI buffer: 2ul
- H2O: 6ul
- Total: 20ul
- Enzyme digestion: D-term E with XbaI
- D-term E: 30ul
- XbaI: 2ul
- 10x 2 buffer: 5ul
- 10x BSA: 5ul
- H2O: 8ul
- Total: 50ul
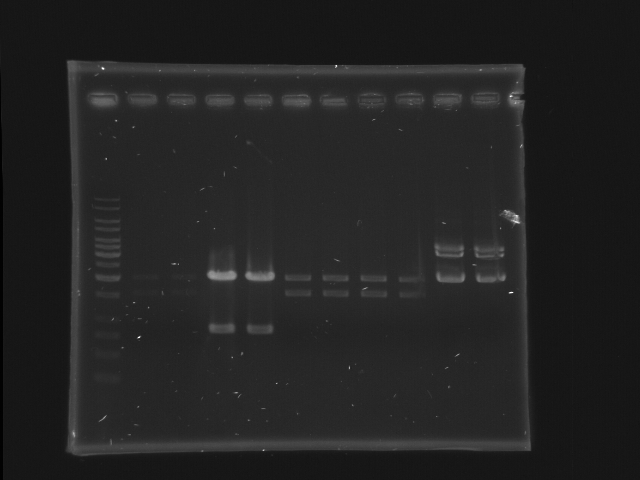
Gel separation
- 1% gel, TAE 1X, 30 min, 100v
- 11 lanes (left most is marker)
- each sample (24 uL) are separated into 2 lanes (12 uL for each lane)
- CinR+HSL #1-#4 (lane 2-9) and D-term (lane 10-11)
- use 1Kb ladder
- CinR+HSL insert size = 1.53 Kb
- D-term vector size = 3.284 Kb
- 6X dye
- total volume after digestion is 20uL
- thus, the dye is 4uL
- X/(20+X) = 1/6, X = 4
- sample after dye addition is 20 + 4 = 24 uL
- Lane 1: 1kb ladder (5ul)
- Lane 2, 3: CinR+HSL 1 EcoRI, SpeI
- Lane 4, 5: CinR+HSL 2 EcoRI, SpeI
- Lane 6, 7: CinR+HSL 3 EcoRI, SpeI
- Lane 8, 9: CinR+HSL 4 EcoRI, SpeI
- Lane 10, 11: D-term EcoRI
- Comments
- The concentration of CinR+HSL 1 is too low.
- The insert of CinR+HSL 2 is wrong.
- EcoRI digestion of D-term is incomplete, hence only the upper band is isolated for DNA extraction. (The lower band is probably the negative supercoiled form of the plasmid)