UCSF/Cloning Strategy1
From 2007.igem.org
| Line 1: | Line 1: | ||
| - | 1- Cloning Strategy | + | '''1- Cloning Strategy |
a- Multipart Cloning of Plasmid-Encoded Promoters, Negative Effectors and Recruitment Domains | a- Multipart Cloning of Plasmid-Encoded Promoters, Negative Effectors and Recruitment Domains | ||
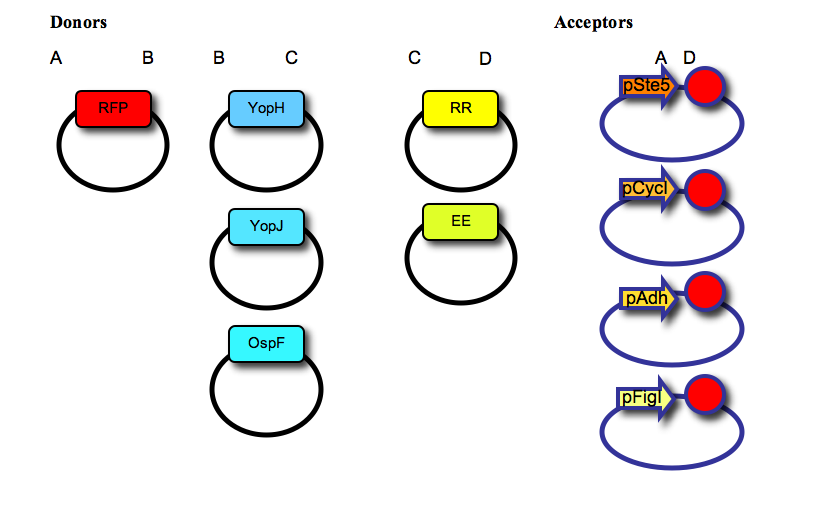
| - | We have used a multipart cloning strategy based on the Type IIs restriction enzyme AarI to generate, in a single ligation, a vector containing: a promoter, a fluorescent protein tag, a negative effector, a recruitment domain (synthetic leucine zipper) and a transcription terminator. For that, we first made two families of “Donor” and “Acceptor” vectors, based on the yeast expression vector pRS315. The 5 donor vectors contain a single part (RFP tag; negative effectors YopH, YopJ or OspF; synthetic leucine zippers for recruitment, “EE” or “RR”) cloned between two AarI sites, with distinct 4-base overhangs (named A, B, C, or D, as shown in the Figure below). The 4 acceptor vectors contain a promoter (pSte5, pCyc, pAdh, or pFig; the first three are constitutive with low, medium or high expression levels respectively, the last one is induced by activation of the mating pathway), two AarI restriction sites (each one with a distinct 4-base overhang, named A and D), and a transcription terminator (red circle); as shown in the figure below: | + | We have used a multipart cloning strategy based on the Type IIs restriction enzyme AarI to generate, in a single ligation, a vector containing: a promoter, a fluorescent protein tag, a negative effector, a recruitment domain (synthetic leucine zipper) and a transcription terminator. For that, we first made two families of “Donor” and “Acceptor” vectors, based on the yeast expression vector pRS315. The 5 donor vectors contain a single part (RFP tag; negative effectors YopH, YopJ or OspF; synthetic leucine zippers for recruitment, “EE” or “RR”) cloned between two AarI sites, with distinct 4-base overhangs (named A, B, C, or D, as shown in the Figure below). The 4 acceptor vectors contain a promoter (pSte5, pCyc, pAdh, or pFig; the first three are constitutive with low, medium or high expression levels respectively, the last one is induced by activation of the mating pathway), two AarI restriction sites (each one with a distinct 4-base overhang, named A and D), and a transcription terminator (red circle); as shown in the figure below:''' |
| - | [[Image: | + | [[Image:DonorAcceptors.png]] |
| - | + | ||
| - | + | ||
| - | + | '''Each final construct was then made by ligation into a given acceptor vector, an RFP tag, followed by a negative effector and a leucine zipper, resulting in the following constructs (one example shown in the Figure):''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 20:20, 3 October 2007
1- Cloning Strategy
a- Multipart Cloning of Plasmid-Encoded Promoters, Negative Effectors and Recruitment Domains
We have used a multipart cloning strategy based on the Type IIs restriction enzyme AarI to generate, in a single ligation, a vector containing: a promoter, a fluorescent protein tag, a negative effector, a recruitment domain (synthetic leucine zipper) and a transcription terminator. For that, we first made two families of “Donor” and “Acceptor” vectors, based on the yeast expression vector pRS315. The 5 donor vectors contain a single part (RFP tag; negative effectors YopH, YopJ or OspF; synthetic leucine zippers for recruitment, “EE” or “RR”) cloned between two AarI sites, with distinct 4-base overhangs (named A, B, C, or D, as shown in the Figure below). The 4 acceptor vectors contain a promoter (pSte5, pCyc, pAdh, or pFig; the first three are constitutive with low, medium or high expression levels respectively, the last one is induced by activation of the mating pathway), two AarI restriction sites (each one with a distinct 4-base overhang, named A and D), and a transcription terminator (red circle); as shown in the figure below:
Each final construct was then made by ligation into a given acceptor vector, an RFP tag, followed by a negative effector and a leucine zipper, resulting in the following constructs (one example shown in the Figure):