ETHZ/Biology/Lab
From 2007.igem.org
m (Added references, removed parts (are shown in biology page)) |
m (Fixed headings, added references.) |
||
| Line 9: | Line 9: | ||
Todo: decide what happens with lab book ([https://2007.igem.org/ETHZ/Lab_book here]) | Todo: decide what happens with lab book ([https://2007.igem.org/ETHZ/Lab_book here]) | ||
| - | == | + | ==Cloning plan== |
| - | === | + | ===Parts assignment into plasmids=== |
Three plasmids are used for the EducatETH <i>E.coli</i> system parts as follows: | Three plasmids are used for the EducatETH <i>E.coli</i> system parts as follows: | ||
| Line 33: | Line 33: | ||
It is important to insert parts responsible for the production of fluorescent proteins in low copy plasmids, as they are potentially harmful for the cell. Unfortunately, working with low copy plasmids makes the procedure more demanding in the lab. | It is important to insert parts responsible for the production of fluorescent proteins in low copy plasmids, as they are potentially harmful for the cell. Unfortunately, working with low copy plasmids makes the procedure more demanding in the lab. | ||
| - | === | + | ===Linkers=== |
Because the plasmids used were not standard plasmids found in the registry, but came from the lab where we work, linkers compatible with the standard BioBrick assembly have to be used in order to work with them. The list of all linkers is the following: | Because the plasmids used were not standard plasmids found in the registry, but came from the lab where we work, linkers compatible with the standard BioBrick assembly have to be used in order to work with them. The list of all linkers is the following: | ||
| Line 65: | Line 65: | ||
Note that four linkers are tested for pbr322, as two are used for the tetracycline-resistance version of pbr322 and two are used for the ampicillin-resistnace version. | Note that four linkers are tested for pbr322, as two are used for the tetracycline-resistance version of pbr322 and two are used for the ampicillin-resistnace version. | ||
| - | === | + | === Procedure === |
The standard BioBrick assembly will be used to put the parts in the plasmids. Detailed information on how the BioBrick part fabrication works can be found [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication here]. For a shorter explanation of how to assemble 2 parts together check [http://partsregistry.org/Assembly:Standard_assembly here]. [[Image:Assembly _process.png|thumb|300px|DNA assembly process ([1]) '''(Fig. 4)''']] Note that the composite part is constructed from the end to the beginning, i.e. each new part is inserted ''before'' the existing one. In the following, the plasmid containing the new part to be inserted will be referred to as the ''donor'' and the plasmid accepting the new part will be referred to as the ''acceptor''. Composite pars made of parts '''a''' and '''b''' are denoted '''a.b'''. | The standard BioBrick assembly will be used to put the parts in the plasmids. Detailed information on how the BioBrick part fabrication works can be found [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication here]. For a shorter explanation of how to assemble 2 parts together check [http://partsregistry.org/Assembly:Standard_assembly here]. [[Image:Assembly _process.png|thumb|300px|DNA assembly process ([1]) '''(Fig. 4)''']] Note that the composite part is constructed from the end to the beginning, i.e. each new part is inserted ''before'' the existing one. In the following, the plasmid containing the new part to be inserted will be referred to as the ''donor'' and the plasmid accepting the new part will be referred to as the ''acceptor''. Composite pars made of parts '''a''' and '''b''' are denoted '''a.b'''. | ||
| - | ==== | + | ==== Plasmid 1 ''(pbr322ap)'' ==== |
# Put parts 1,2,3 in pbr322ap plasmids. | # Put parts 1,2,3 in pbr322ap plasmids. | ||
| Line 75: | Line 75: | ||
# Merge plasmid containing part '''1''' ''(donor)'' with plasmid containing composite part '''2.3''' ''(acceptor)''. You should get a plasmid containing a '''1.2.3''' composite part. | # Merge plasmid containing part '''1''' ''(donor)'' with plasmid containing composite part '''2.3''' ''(acceptor)''. You should get a plasmid containing a '''1.2.3''' composite part. | ||
| - | ==== | + | ==== Plasmid 2 ''(pck01cm)''==== |
# Put parts 4,5,8,9 in pck01cm plasmids. | # Put parts 4,5,8,9 in pck01cm plasmids. | ||
| Line 82: | Line 82: | ||
# Merge plasmid containing composite part '''4.5''' ''(donor)'' with plasmid containing composite part '''8.9''' ''(acceptor)''. You should get a plasmid containing a '''4.5.8.9''' composite part. | # Merge plasmid containing composite part '''4.5''' ''(donor)'' with plasmid containing composite part '''8.9''' ''(acceptor)''. You should get a plasmid containing a '''4.5.8.9''' composite part. | ||
| - | ==== | + | ====Plasmid 3 ''(pacyc177km)''==== |
# Put parts 6,7,10,11 in pacyc177km plasmids. | # Put parts 6,7,10,11 in pacyc177km plasmids. | ||
| Line 89: | Line 89: | ||
# Merge plasmid containing composite part '''6.7''' ''(donor)'' with plasmid containing composite part '''10.11''' ''(acceptor)''. You should get a plasmid containing a '''6.7.10.11''' composite part. | # Merge plasmid containing composite part '''6.7''' ''(donor)'' with plasmid containing composite part '''10.11''' ''(acceptor)''. You should get a plasmid containing a '''6.7.10.11''' composite part. | ||
| - | == | + | == Problems we faced== |
| - | == | + | == Lab book == |
| - | ==== | + | ==== Week 1 ==== |
{| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| Line 146: | Line 146: | ||
|} | |} | ||
| - | ==== | + | ==== Week 2 ==== |
{| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| Line 215: | Line 215: | ||
|} | |} | ||
| - | ==== | + | ==== Week 3 ==== |
Little rearrangements of the parts. Planning of the sequences to order them. | Little rearrangements of the parts. Planning of the sequences to order them. | ||
| - | ==== | + | ==== Week 4 ==== |
{| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| Line 270: | Line 270: | ||
|} | |} | ||
| - | ==== | + | ==== Week 5 ==== |
{| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| Line 340: | Line 340: | ||
|} | |} | ||
| - | ==== | + | ==== Week 6 ==== |
{| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| Line 407: | Line 407: | ||
|} | |} | ||
| - | ==== | + | ==== Week 7 ==== |
{| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| Line 481: | Line 481: | ||
|} | |} | ||
| - | ==== | + | ==== Week 8 ==== |
{| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| Line 525: | Line 525: | ||
|} | |} | ||
| - | ==== | + | ====Week 9 ==== |
{| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| Line 569: | Line 569: | ||
|} | |} | ||
| - | ==== | + | ==== Week 10 ==== |
{| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
Revision as of 08:22, 20 October 2007

In this page, you can find information on laboratory conducted to construct EducatETH E.coli. The system parts are presented again, their assembly into plasmids and the cloning plan are explained and all lab notes taken by the ETH Zurich team are accessible. If you are trying to construct EducatETH E.coli at your lab, the section Problems we faced might be useful to you. If you want to see the whole biological design of the system, please visit the Biology Pespective. Finally, photos of our lab experience are accessible under Pictures!
Todo: decide what happens with lab book (here)
Contents |
Cloning plan
Parts assignment into plasmids
Three plasmids are used for the EducatETH E.coli system parts as follows:
| plasmid | resistance | copy type | contents | comments |
|---|---|---|---|---|
| pbr322 | ampicillin | high | 1,2,3 | constitutive subsystem |
| pck01 | chloramphenicol | low | 4,5,8,9 | reporting subsystem |
| pacyc177 | kanamycin | low | 6,7,10,11 | learning subsystem, reporting subsystem |
It is important to insert parts responsible for the production of fluorescent proteins in low copy plasmids, as they are potentially harmful for the cell. Unfortunately, working with low copy plasmids makes the procedure more demanding in the lab.
Linkers
Because the plasmids used were not standard plasmids found in the registry, but came from the lab where we work, linkers compatible with the standard BioBrick assembly have to be used in order to work with them. The list of all linkers is the following:
| Linker | Plasmid |
|---|---|
| pbr322-1 | pbr322 |
| pbr322-2 | pbr322 |
| pbr322-3 | pbr322 |
| pbr322-4 | pbr322 |
| pck01 | pck01 |
| pck01-2 | pck01 |
| pacyc177-1 | pacyc177 |
| pacyc177-2 | pacyc177 |
Note that four linkers are tested for pbr322, as two are used for the tetracycline-resistance version of pbr322 and two are used for the ampicillin-resistnace version.
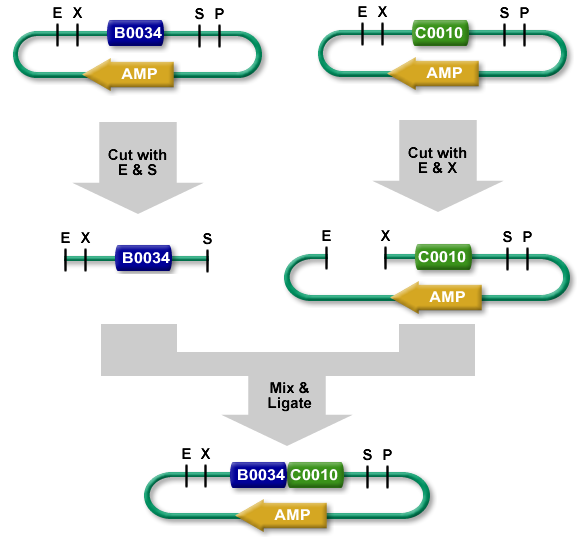
Procedure
The standard BioBrick assembly will be used to put the parts in the plasmids. Detailed information on how the BioBrick part fabrication works can be found [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication here]. For a shorter explanation of how to assemble 2 parts together check [http://partsregistry.org/Assembly:Standard_assembly here]. Note that the composite part is constructed from the end to the beginning, i.e. each new part is inserted before the existing one. In the following, the plasmid containing the new part to be inserted will be referred to as the donor and the plasmid accepting the new part will be referred to as the acceptor. Composite pars made of parts a and b are denoted a.b.Plasmid 1 (pbr322ap)
- Put parts 1,2,3 in pbr322ap plasmids.
- Merge plasmid containing part 2 (donor) with plasmid containing part 3 (acceptor). You should get a plasmid containing a 2.3 composite part.
- Merge plasmid containing part 1 (donor) with plasmid containing composite part 2.3 (acceptor). You should get a plasmid containing a 1.2.3 composite part.
Plasmid 2 (pck01cm)
- Put parts 4,5,8,9 in pck01cm plasmids.
- Merge plasmid containing part 4 (donor) with plasmid containing part 5 (acceptor). You should get a plasmid containing a 4.5 composite part.
- Merge plasmid containing part 8 (donor) with plasmid containing part 9 (acceptor). You should get a plasmid containing a 8.9 composite part. Note: this step can be done simultaneously with the above.
- Merge plasmid containing composite part 4.5 (donor) with plasmid containing composite part 8.9 (acceptor). You should get a plasmid containing a 4.5.8.9 composite part.
Plasmid 3 (pacyc177km)
- Put parts 6,7,10,11 in pacyc177km plasmids.
- Merge plasmid containing part 6 (donor) with plasmid containing part 7 (acceptor). You should get a plasmid containing a 6.7 composite part.
- Merge plasmid containing part 10 (donor) with plasmid containing part 11 (acceptor). You should get a plasmid containing a 10.11 composite part. Note: this step can be done simultaneously with the above.
- Merge plasmid containing composite part 6.7 (donor) with plasmid containing composite part 10.11 (acceptor). You should get a plasmid containing a 6.7.10.11 composite part.
Problems we faced
Lab book
Week 1
| Date | TODO's | Completed | People |
|---|---|---|---|
| Mon, 06. Aug. 2007 |
| Sylke Raphael Stefan Markus Martin Christos Joe | |
| Tue, 07. Aug. 2007 |
| Sylke Raphael Stefan Markus Martin Christos Joe | |
| Wed, 08. Aug. 2007 |
| Raphael Stefan | |
| Thu, 09. Aug. 2007 |
| Raphael Stefan Martin Christos Joe | |
| Fri, 10. Aug. 2007 |
|
Christos | |
| Sat, 11. Aug. 2007 | no labwork | ||
| Sun, 12. Aug. 2007 | labwork cancelled |
Week 2
| Date | TODO's | Completed | People |
|---|---|---|---|
| Mon, 13. Aug. 2007 start at 3 pm |
|
| Martin Markus Christos Tim |
| Tue, 14. Aug. 2007 | Morning Shift:
Evening Shift:
| Morning Shift:
Evening Shift:
| Morning Shift (9am-1pm?): Markus Tim Evening Shift (5pm-...): Martin Christos |
| Wed, 15. Aug. 2007 |
|
| From 12: Martin Markus |
| Thu, 16. Aug. 2007 |
|
|
Markus |
| Fri, 17. Aug. 2007 |
|
| Martin |
| Sat, 18. Aug. 2007 | |||
| Sun, 19. Aug. 2007 |
Week 3
Little rearrangements of the parts. Planning of the sequences to order them.
Week 4
| Date | TODO's | Completed | People |
|---|---|---|---|
| Mon, 27. Aug. 2007 | |||
| Tue, 28. Aug. 2007 | |||
| Wed, 29. Aug. 2007 | |||
| Thu, 30. Aug. 2007 | |||
| Fri, 31. Aug. 2007 | |||
| Sat, 01. Sept. 2007 |
|
| Stefan |
| Sun, 02. Sept. 2007 |
|
| Stefan |
Week 5
| Date | TODO's | Completed | People |
|---|---|---|---|
| Mon, 03. Sept. 2007 |
|
| Martin Stefan |
| Tue, 04. Sept. 2007 |
|
| Martin Christian |
| Wed, 05. Sept. 2007 |
|
| Martin |
| Thu, 06. Sept. 2007 |
|
| Christian |
| Fri, 07. Sept. 2007 |
|
| Martin |
| Sat, 08. Sept. 2007 | |||
| Sun, 09. Sept. 2007 |
Week 6
| Date | TODO's | Completed | People |
|---|---|---|---|
| Mon, 10. Sept. 2007 |
all digests o/n |
Christian | |
| Tue, 11. Sept. 2007 |
|
|
Christian |
| Wed, 12. Sept. 2007 |
|
Christian | |
| Thu, 13. Sept. 2007 |
|
Christian | |
| Fri, 14. Sept. 2007 |
|
*pBR322-MCS (Tet-selection) clone2 positive |
Christian |
| Sat, 15. Sept. 2007 | |||
| Sun, 16. Sept. 2007 |
Week 7
| Date | TODO's | Completed | People |
|---|---|---|---|
| Mon, 17. Sept. 2007 |
|
|
Christian |
| Tue, 18. Sept. 2007 |
|
> no DNA on pACYC177 digest-gel, only degradation smear
|
Christian |
| Wed, 19. Sept. 2007 |
| ||
| Thu, 20. Sept. 2007 | |||
| Fri, 21. Sept. 2007 | |||
| Sat, 22. Sept. 2007 | |||
| Sun, 23. Sept. 2007 |
Week 8
| Date | TODO's | Completed | People |
|---|---|---|---|
| Mon, 24. Sept. 2007 | |||
| Tue, 25. Sept. 2007 | |||
| Wed, 26. Sept. 2007 | |||
| Thu, 27. Sept. 2007 | |||
| Fri, 28. Sept. 2007 | |||
| Sat, 29. Sept. 2007 | |||
| Sun, 30. Sept. 2007 |
Week 9
| Date | TODO's | Completed | People |
|---|---|---|---|
| Mon, 1. Oct. 2007 | |||
| Tue, 2. Oct. 2007 | |||
| Wed, 3. Oct. 2007 | |||
| Thu, 4. Oct. 2007 | |||
| Fri, 5. Oct. 2007 | |||
| Sat, 6. Oct. 2007 | |||
| Sun, 7. Oct. 2007 |
Week 10
| Date | TODO's | Completed | People |
|---|---|---|---|
| Mon, 8. Oct. 2007 | |||
| Tue, 9. Oct. 2007 | |||
| Wed, 10. Oct. 2007 | |||
| Thu, 11. Oct. 2007 | |||
| Fri, 12. Oct. 2007 | |||
| Sat, 13. Oct. 2007 | |||
| Sun, 14. Oct. 2007 |
References
[1] [http://partsregistry.org/Assembly:Standard_assembly Standard Assembly Process]