Davidson Missouri W/Gene splitting
From 2007.igem.org
(→The Genes) |
(→A Closer Look) |
||
| Line 38: | Line 38: | ||
=A Closer Look= | =A Closer Look= | ||
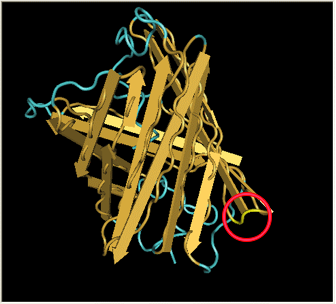
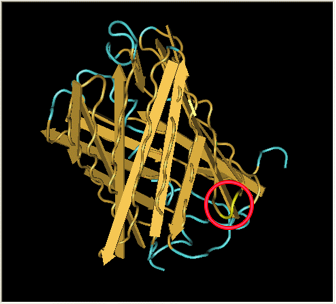
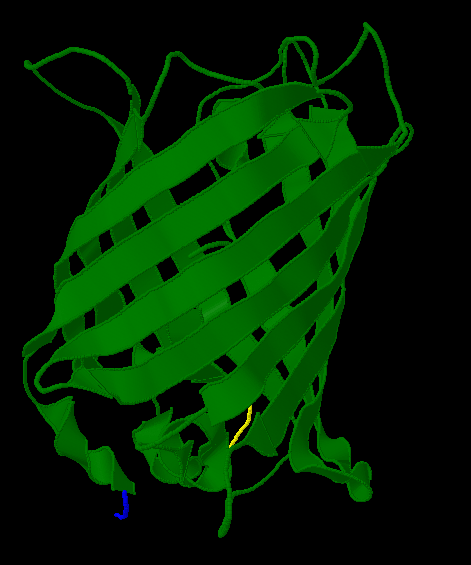
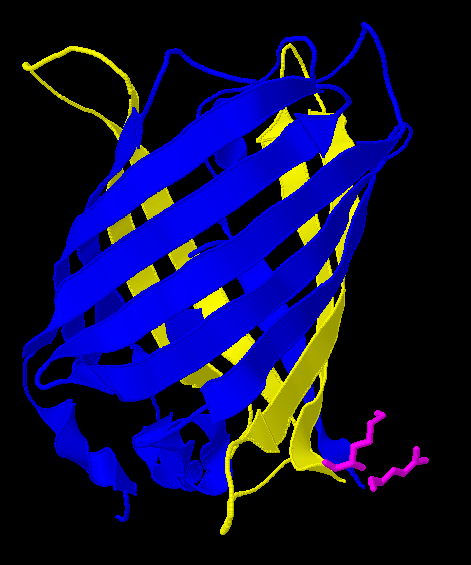
| - | Here is a more detailed view of splitting GFP. | + | Here is a more detailed view of splitting GFP. The first image, with the protein structure highlighted in green, shows the normal, wild-type structure of the protein. The second image highlights the two parts of the protein on either side of the ''hixC'' insertion, shown in pink. The additional 13 amino acids are not shown, but they would extend from one pink amino acid into the next. |
| - | [[Image:Gfp-nosplit.png|thumb| | + | [[Image:Gfp-nosplit.png|thumb|340px|none|]][[Image:Gfp-split.png|thumb|340px|none|]] |
Revision as of 23:24, 20 October 2007
What is Gene Splitting?
"Gene splitting" refers to the insertion of a hixC site within the coding region of a gene. Although this allows us to create edges for the simulation of a graph, it will change the protein sequence, potentially interfering with proper functionality. We successfully inserted hixC in two different reporter genes, GFP and RFP. Cells transformed with plasmids containing these "split" genes still fluoresce the appropriate color.
To facilitate the splitting process we developed software to help us. Our [http://gcat.davidson.edu/iGEM07/genesplitter.html online] gene splitting web tool (click here for a tutorial) helps us choose PCR primers that will amplify the appropriate segments of a gene of interest.
The Genes
A Closer Look
Here is a more detailed view of splitting GFP. The first image, with the protein structure highlighted in green, shows the normal, wild-type structure of the protein. The second image highlights the two parts of the protein on either side of the hixC insertion, shown in pink. The additional 13 amino acids are not shown, but they would extend from one pink amino acid into the next.