Tristable
From 2007.igem.org
(→AraC/BAD) |
(→Tri-stable Toggle Switch) |
||
| Line 69: | Line 69: | ||
===Tri-stable Toggle Switch=== | ===Tri-stable Toggle Switch=== | ||
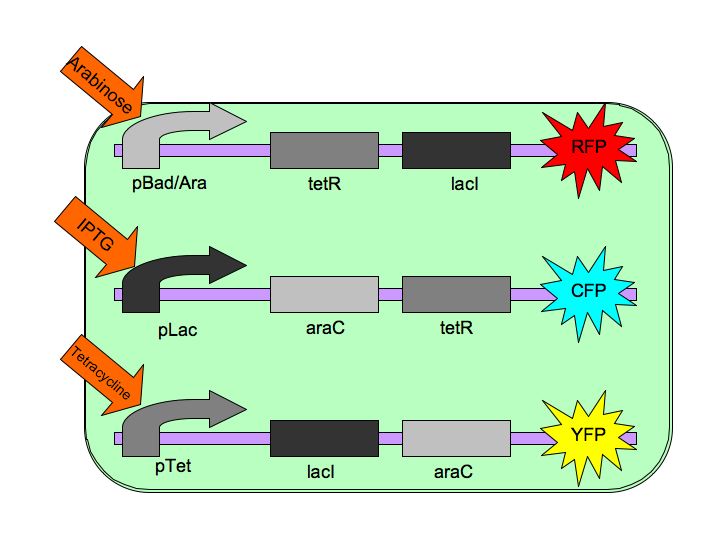
| - | The purpose of the Tri-stable Toggle Switch is to produce three distinct, continuous, and stable outputs in response to three distinct inputs. These three inputs are three separate chemicals which will each induce one state of the switch. [[Image:Tristable_Toggle_Switch_2007.jpg|thumb|left|The Tri-stable Toggle Switch Architecture]] In order to achieve this goal, we are constructing three constructs, each of which consists of a repressible, constitutively-on promoter attached to two repressors. Specifically, our three constructs are pBAD->LacI->TetR, pLacI->AraC->TetR, and pTet->AraC->LacI, where | + | The purpose of the Tri-stable Toggle Switch is to produce three distinct, continuous, and stable outputs in response to three distinct inputs. These three inputs are three separate chemicals which will each induce one state of the switch. [[Image:Tristable_Toggle_Switch_2007.jpg|thumb|left|The Tri-stable Toggle Switch Architecture]] In order to achieve this goal, we are constructing three constructs, each of which consists of a repressible, constitutively-on promoter attached to two repressors. Specifically, our three constructs are pBAD->LacI->TetR, pLacI->AraC->TetR, and pTet->AraC->LacI, where AraC represses pAraC/BAD, [http://en.wikipedia.org/wiki/Lac_repressor LacI] represses pLac and [http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation TetR] represses pTet. Each of the three repressors are inactivated by one of three chemicals, the three inducer chemicals mentioned earlier. These three([http://en.wikipedia.org/wiki/Arabinose arabinose], [http://en.wikipedia.org/wiki/IPTG IPTG] (Isopropyl β-D-1-thiogalactopyranoside) and [http://en.wikipedia.org/wiki/Tetracycline Tetracycline], respectively), cause conformational changes in their respective repressor proteins which keeps them from binding to DNA in an inhibitory manner which leads to gene expression. For example, in the presence of arabinose, AraC cannot repress pBAD so LacI and TetR are produced which in turn repress pTet and pLac and the pAraC/BAD construct is turned on. |
| Line 76: | Line 76: | ||
====AraC/BAD==== | ====AraC/BAD==== | ||
| - | The gene AraC one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html] | + | The gene AraC, one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html] |
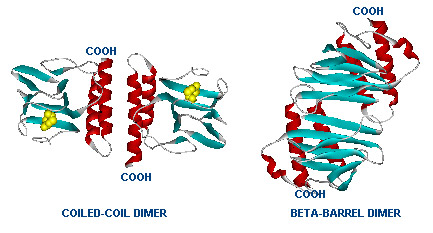
[[Image:Two_Dimers_of_AraC.jpg|thumb|left|Dimer structure with arabinose on the left (yellow)]] | [[Image:Two_Dimers_of_AraC.jpg|thumb|left|Dimer structure with arabinose on the left (yellow)]] | ||
| - | [[Image:AraC_Promoters.gif|left|thumb|The left image shows the araC dimer repressing transcription, while the right conformation enables transcription]]The protein forms a dimer | + | [[Image:AraC_Promoters.gif|left|thumb|The left image shows the araC dimer repressing transcription, while the right conformation enables transcription]]The protein forms a dimer with and without arabinose but the structural change activates or represses the pAraC/BAD. |
| + | |||
| + | |||
| + | |||
====LacI==== | ====LacI==== | ||
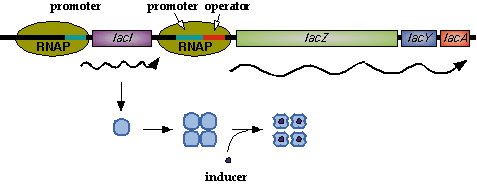
| - | In nature, LacI represses pLac which promotes LacYZA genes that metabolize lactose | + | In nature, LacI represses pLac which promotes the LacYZA genes that metabolize lactose. Thus LacI represses pLac except in the presence of lactose (or lactose mimics, eg IPTG). [[Image:LacI_repressor.gif|thumb|left|Image[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html]. LacI forms a tetramer and represses pLac. However, an inducer, such as IPTG, causes a conformation change that removes LacI from the operator site.]] Lactose causes a conformational change which inhibits LacI from binding to the operator site of pLac. Four LacI proteins form a tetramer to inhibit pLac and four inducer molecules are required to cause the full conformational change in the inhibitor.[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html] |
| Line 93: | Line 96: | ||
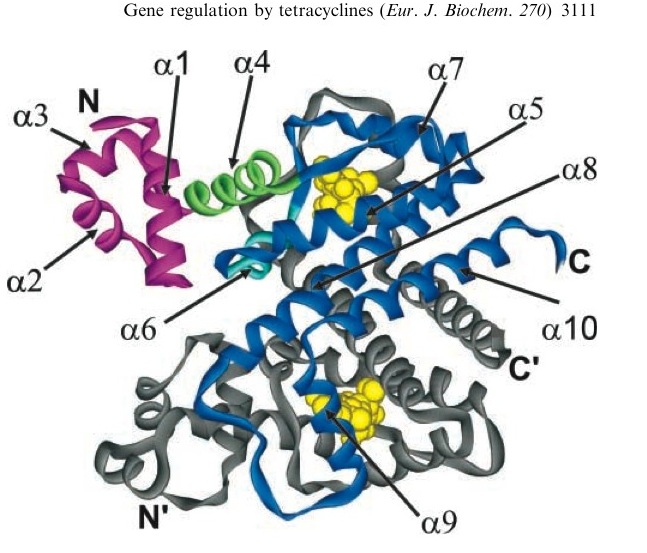
[[Image:Tc_bound_to_TetR.jpg|thumb|left|A tetracycline molecule binds to each of the two TetR monomers to form a dimer]] | [[Image:Tc_bound_to_TetR.jpg|thumb|left|A tetracycline molecule binds to each of the two TetR monomers to form a dimer]] | ||
Tetracycline is highly diffusable through cell membrane (permeation coeficient or 5.6±1.9 * 10^-9 cm/s or half equilibrium time = 35 ± 15 min) and TetR shows a very high affinity for the molecule. The binding constant of TetR to [tc-Mg+] is Ka ~ 10^9 M^-1. When bound to tc, TetR has a low binding level to DNA of 10^5 M^-1. [http://content.febsjournal.org/cgi/content/abstract/270/15/3109] | Tetracycline is highly diffusable through cell membrane (permeation coeficient or 5.6±1.9 * 10^-9 cm/s or half equilibrium time = 35 ± 15 min) and TetR shows a very high affinity for the molecule. The binding constant of TetR to [tc-Mg+] is Ka ~ 10^9 M^-1. When bound to tc, TetR has a low binding level to DNA of 10^5 M^-1. [http://content.febsjournal.org/cgi/content/abstract/270/15/3109] | ||
| - | |||
| - | |||
===Modeling=== | ===Modeling=== | ||
Revision as of 01:03, 24 October 2007

|
||||||||||||||||||