Brown
From 2007.igem.org
(→Modeling) |
|||
| Line 97: | Line 97: | ||
[http://parts2.mit.edu/wiki/index.php/Table_of_preliminary_model_constants Initial Table of Constants] | [http://parts2.mit.edu/wiki/index.php/Table_of_preliminary_model_constants Initial Table of Constants] | ||
| + | |||
| + | [http://parts2.mit.edu/wiki/index.php/Derivation_of_the_Model_Equations Derivation of Model Equations] | ||
Revision as of 04:39, 20 June 2007
Contents |
Welcome to our World
Projects
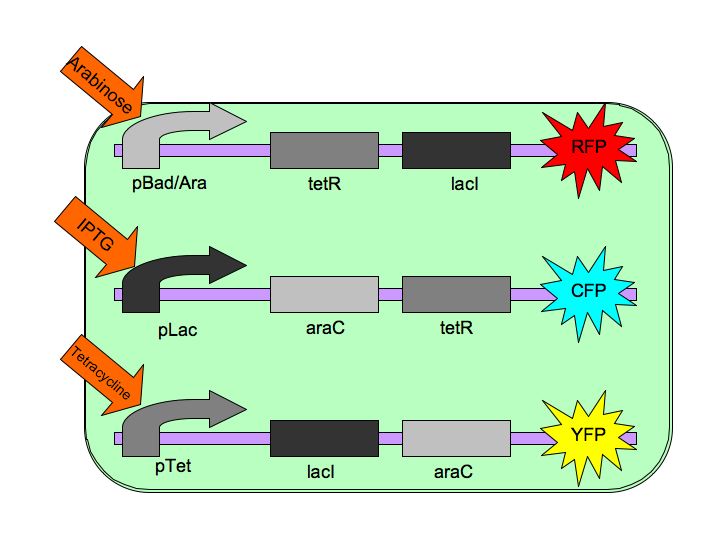
Tri-stable Toggle Switch
The Tri-stable Toggle Switch will be able to produce three distinct, continuous (stable) outputs for each of the three inputs. A chemical will induce the system to "lock into" one state while repressing the other two states. Our three constructs are pBAD->LacI->TetR, pLacI->AraC->TetR, and pTet->AraC->LacI, where [http://en.wikipedia.org/wiki/Bcl-2-associated_death_promoter AraC] represses pBAD, [http://en.wikipedia.org/wiki/Lac_repressor LacI] represses pLac and [http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation TetR] represses pTet. The three chemicals ([http://en.wikipedia.org/wiki/Arabinose arabinose], [http://en.wikipedia.org/wiki/IPTG IPTG] (Isopropyl β-D-1-thiogalactopyranoside) and [http://en.wikipedia.org/wiki/Tetracycline Tetracycline], respectively), cause conformational changes in their respective repressor proteins which leads to gene expression. For example, in the presence of arabinose, AraC cannot repress pBAD so LacI and TetR are produced which in turn repress pTet and pLac.
AraC/BAD
The gene AraC one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html]
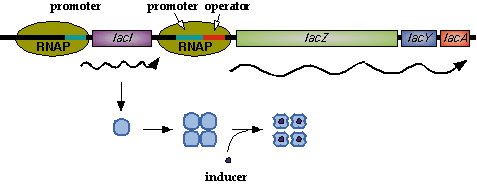
LacI
In nature, LacI represses pLac which promotes LacYZA genes that metabolize lactose, thus LacI represses pLac except in the presence of lactose (or lactose mimics, eg IPTG).
TetR
TetR represses the constitutive promoter pTet. In the presence of tetracycline, an antibiotic, a conformational change in TetR inhibits the protein from binding to the operator region. In nature, pTet promotes TetR and TetA. The latter which acts to pump tetracycline out of the cell, thus the pump is only activated in the presence of Tetracycline.[http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation] The TetR, as it turns out is a very tight repressor and a range of 0 to 1 ug/ml has been shown to cause a 5 order of magnitude change in luciferase production.[http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1319065&query_hl=1&itool=pubmed_docsum]
Tetracycline is highly diffusable through cell membrane (permeation coeficient or 5.6±1.9 * 10^-9 cm/s or half equilibrium time = 35 ± 15 min) and TetR shows a very high affinity for the molecule. The binding constant of TetR to [tc-Mg+] is Ka ~ 10^9 M^-1. When bound to tc, TetR has a low binding level to DNA of 10^5 M^-1. [http://content.febsjournal.org/cgi/content/abstract/270/15/3109]
Modeling
Brown iGEM 2006 Matlab model code Media:tristable2006.txt
[http://parts2.mit.edu/wiki/index.php/Table_of_preliminary_model_constants Initial Table of Constants]
[http://parts2.mit.edu/wiki/index.php/Derivation_of_the_Model_Equations Derivation of Model Equations]