Lead
From 2007.igem.org
(Difference between revisions)
| Line 229: | Line 229: | ||
#The cooperativity of repression, Beta. | #The cooperativity of repression, Beta. | ||
| - | In the Model, the repressor | + | In the Model, the repressor strength = [Repressor]^Beta |
| + | |||
| + | The Beta value is characteristic of each repressor and represents a constant that cannot be changed. | ||
| + | |||
| + | ===Degradation=== | ||
| + | The degradation in the model is denoted by the term "-repressor," where the degradation constant is unity because the rates are relative, rather than absolute. The next iteration of the model will include degradation constants. Currently in the registry all of the repressors have strong degradation tags. Jason Kelly made a comment on one repressor and said he was suspicious of how strong of a repression would be able to be obtained from the repessors. We need strong repression for our system, so we may need to remove the tag our system to function. | ||
| + | |||
| + | ==The Model== | ||
| + | [[Image:model equation.png|Tri-Stable Mathematical Model]] | ||
| + | |||
| + | |||
| + | *Beta = Repression Strength | ||
| + | *Alpha = Repressor Production Rate | ||
| + | *X = Repressor1 | ||
| + | *Y = Repressor2 | ||
| + | *Z = Repressor3 | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===Stability=== | ||
| + | [[Image:tri stableRegion.png|left|thumb|Tri-Stable region solved in Matlab]] | ||
| + | |||
| + | Blue points are stable combinations of repressor production rates, while red circles are unstable combinations of repressor production rates. This graph is for Beta = 2. The tristable region gets larger (i.e. more disparate alpha values will be able to constitute a tristable system) as beta gets larger. The tristable region disappears when Beta equals one or less. | ||
| + | |||
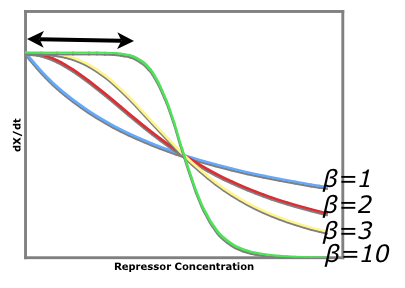
| + | An easy visual aid to seeing how increased beta values (cooperativity of repression) leads to a more stable system consider the following graph of rate of production of repressor1 vs. the repressor that inhibits the production of repressor1. dx/dt vs. [y] | ||
| + | |||
| + | [[Image:beta values.png|Comparison of different beta values]] | ||
| + | |||
| + | The black arrow shows that there is a larger region for larger beta values where stray repessors will not significantly affect the rate of repressor production rate whereas in a system of beta = 1, stray repressors will significantly change the system, making it less robust. | ||
| + | |||
| + | =Testing Constructs= | ||
| + | |||
| + | There are two methods we could follow in designing the Switch. We could randomly try different RBSs, hope it works and if not try again without having much of an understanding of why our cnostructs didn't work. Or we can test our repressors, promoters and inducers and have a systematic approach to anaylizing our system so that when something works or doesn't work we will know why. Thus we have designed a few tests which should give us relative and absolute values of our system that we can then plug into the model. | ||
| + | |||
| + | We decided to to test for three values. The combined transcription/translation rate of each repressor, the cooperativity of each repressor and the concentration of ligand needed to deactivate each repressor. We managed to design three tests all using the same constructs so as to minimize ligations. These tests should determine our variables independantly, i.e. changing synthesis rate should not change cooperativity of repression. | ||
| + | |||
| + | ==Synthesis Rate== | ||
| + | The combined transcription/translation rate of the repressor is the combined strength of the promoter and the RBS. In [[Image:alphaTest.png|thumb|left|The Alpha test Architecture]]our model this is the alpha value. Our model predicts that the alpha values for each repressor should be fairly comparable (the stable region is along the 1 to 1 to 1 line in 3D). Since we can't change the promoter strength very easily, we will change the RBS strength to obtain similar alpha values for all repressors. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
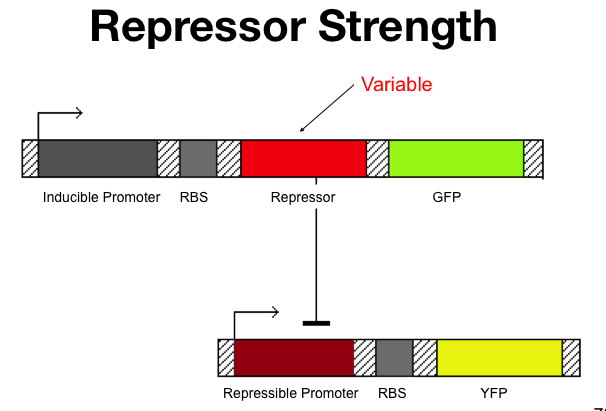
| + | ==Cooperativity of Repression== | ||
| + | The cooperativity describes an inherent characteristic of a repressor's repression. In our system we want to know how [[Image:betaTest.png|thumb|left|The Beta test Architecture]]much increased repressor concentration will increase repression. In our model, cooperativity is an exponent, beta, so that the repressor concentration is raised to the beta ([repressor]^beta = total repression). For beta = 1, repression increases linearly with repressor concentration. With twice as much repressor there is twice the repression. For beta = 2, repression increases with the square of the concetration. Twce as much repressor leads to four times the repression. Our model predicts that our system will be more robust with greater alpha values and the tri stable region (in the graph) will be larger. Beta must be larger than one for the system to be stable. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==Ligand Concentration== | ||
| + | Naturally, we don't want to add more ligand than we need. In what ever application the project might find, if we wanted to change the state again by adding a different [[Image:ligandTest.png|thumb|left|The Ligand test Architecture]]ligand, we wouldn't want excessive amounts of the first ligand floating around. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =Appendix= | ||
| + | ==Schedule of Construction== | ||
| + | We have a schedule of ligations and transformations that we keep on googledocs to easily access information about the dna and cells in our freezer and fridge. This also organizes what ligations we plan to do in the future. In all there are about 40 ligations we want to do to construct the test constructs and first tristable system and based on the results of the tests, we will need to do as many as 20 more ligations. | ||
| + | |||
| + | [[Image:Transformations.png|left|thumb|Brown iGEM transformation database on googledocs]] | ||
| + | |||
| + | [[Image:Ligations.png|thumb|right|Brown iGEM ligation database on googledocs]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==Other Predicted Obstacles== | ||
| + | #Other iGEM teams (info via Patrick King) have found that the araC gene contains a promoter region. Grown in TOP10 cells the construct araC>RBS>GFP>Terminator was observed to give a strong florescent output. In our system, this would give a florescent output for one FP even if that construct was not on. | ||
| + | #AraC exhibits all or none gene expression in the presents of arabinose because the natural transporter, when induced, causes that cell to transport arabinose, depleting the culture. This may be overcome by adding a saturating level of arabinose. In the arabinose construct already made, we were not able to observe FP output even though there was a high level of arabinose. We used XL1Blue cells. | ||
| + | #The degradation tags on the repressors may not give a long enough protein life time to yield strong repression. Thus, the tags may need to be removed to have a working device. | ||
| + | |||
| + | ==Email Correspondances about Tri-Stable Switch== | ||
| + | [[Media:emailwithJasonLohmueller.txt]] | ||
| + | |||
| + | [[Media:emailwithNicolaNeretti.txt]] | ||
| + | |||
| + | [[Media:emailwithPatrickKing.txt]] | ||
| + | --> | ||
| + | <html> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </td> | ||
| + | <td width=255 bgcolor="#e8e8e8" valign=top align=left> | ||
| + | |||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </html> | ||
Revision as of 21:57, 26 October 2007

|
|||||||||||||||||