PennState/Project/Diauxie/PromoterTest
From 2007.igem.org
| Line 1: | Line 1: | ||
'''Promoter Test Constructs''' | '''Promoter Test Constructs''' | ||
| - | To test | + | The xyl promoter region promotes in both the direction of genes encoding for both xylA and xylF metabolization and transport proteins. The CPR-cAMP, and xylR binding sites regulate both directions. This means that there are four possibilities for making promoters: xylF facing including the CRP-cAMP binding site, xylF facing without the CRP-cAMP binding site, xylA facing including the CRP-cAMP binding site, and xylF facing without the CRP-cAMP binding site. |
| + | |||
| + | To test the four xyl promoters, each is placed in a construct followed by a ribosome binding site, the reporter green florescent protein, and a terminator. The strength of each is measured by gfp emission. | ||
Revision as of 23:38, 26 October 2007
Promoter Test Constructs
The xyl promoter region promotes in both the direction of genes encoding for both xylA and xylF metabolization and transport proteins. The CPR-cAMP, and xylR binding sites regulate both directions. This means that there are four possibilities for making promoters: xylF facing including the CRP-cAMP binding site, xylF facing without the CRP-cAMP binding site, xylA facing including the CRP-cAMP binding site, and xylF facing without the CRP-cAMP binding site.
To test the four xyl promoters, each is placed in a construct followed by a ribosome binding site, the reporter green florescent protein, and a terminator. The strength of each is measured by gfp emission.
 This construct uses the xylF and xylA facing promoters that includes the CRP-cAMP binding region.
This construct uses the xylF and xylA facing promoters that includes the CRP-cAMP binding region.
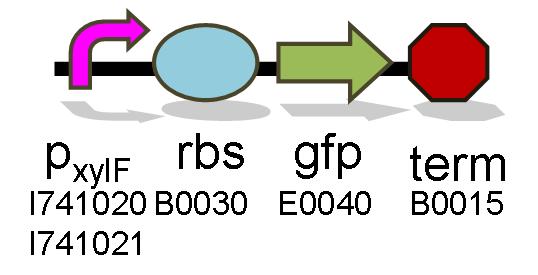 This construct uses the xylF and xylA facing promoters that do not include the CRP-cAMP binding region.
This construct uses the xylF and xylA facing promoters that do not include the CRP-cAMP binding region.
After each of the four promoter constructs are tested individually, the two which include the CRP-cAMP are changed so that a gene expressing the mutant CRP* is included.

This construct uses the xylF and xylA facing promoters that do not include the CRP-cAMP binding region followed by the CRP* gene.