Paris/July 16
From 2007.igem.org
David.bikard (Talk | contribs) (→PCRs) |
Nicolas C. (Talk | contribs) (→Transformations of Biobricks) |
||
| Line 59: | Line 59: | ||
== Transformations of Biobricks == | == Transformations of Biobricks == | ||
| + | As in the protocol given in the registry. | ||
| + | Transformation of biobricks : | ||
| + | * BBa_B00300 pSB1A2 : RBS (well 3G plate 1) | ||
| + | * BBa_E0422 pSB1A2 : ECFP (RBS++Term) (well 11G plate 1) | ||
| + | * BBa_E0241 pSB1A2 : PoPs to GFP converter (well 15c plate 2) | ||
| + | * BBa_E0840 : (well 16E plate 1) | ||
| + | * BBa_J61047 pSB1A2 : Cre ORF (well 8P plate 4) | ||
| + | Transformation in DH5alpha subcloning efficiency | ||
| + | Spread on LB-Amp | ||
Revision as of 12:24, 17 July 2007
Contents |
Plasmid pKs::DGAT expression in E. Coli
We tried to see TG with NR died on E. Coli cells transfected with pKs::DGAT, an IPTG-inducible promoter.
We tried different growth media containing more or less oleate, that should in theory increase TG synthesis.
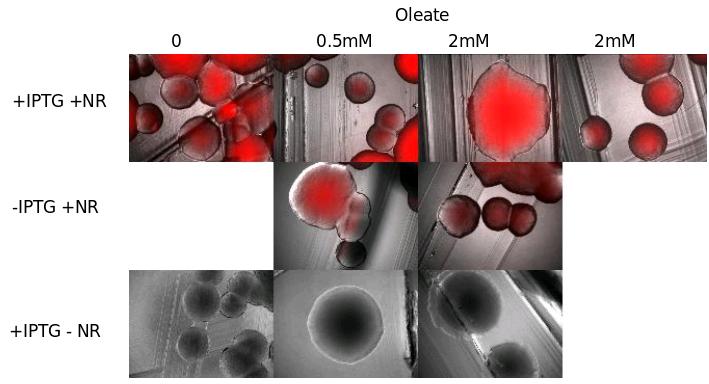
Results : we don't see the inductible effect of IPTG. We can think that :
- Either the fluorescence without IPTG is due to a leak of the promoter
- Either DGAT is not induce in presence of IPTG, and the fluorescence we see is only a background.
Growth kinetics of w121 strain
Results of the previous day : we lost everything because of a crash of the computer :( Sorry Eimad !
Transduction of MG1655 with P1 stock made on w121
- Control (1mL LB MgSO4 30mM; CaCl2 15mM)
- 5µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture ON
- 50µL Phage + 900µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture ON
- 500µL Phage + 500µL LB (MgSO4 30mM; CaCl2 15mM) + 100µL MG1655 Culture ON
=>For the nth time, it is not working : we have only contaminants.
MiniPreps
- I0500 clones 1, 2
- pJ23107 clones 1, 2
PCR purification
- Lox71-FtsZ1
- FtsZ2
- DGAT1
- DGAT2
PCRs
The Lox66-DapAColi PCR did not work... We'll try again with different annealing temperature
Assembly PCRs:
- Lox71-FtsA-FtsZ-1 + FtsZ-2
- DGAT-1 + DGAT-2
The Lox66-DapAColi PCR did not work... so we try a gradient of annealing temperatures
| PCR : Lox66-DapAColi | ||
|---|---|---|
| Name | Lox66-DapAColi | |
| Annealing T° | 50-65°C | |
| Time of elongation | 2m00' | |
| Number of Cycles | 35 | |
| Buffer | 5x 10µL | |
| MgCl2 | 10µM 0µL | |
| dNTP | 10µM 1µL | |
| oligoF | 6 Lox66-DapAColi-F | 10µM 2.5µL |
| oligoR | 7 DapAColi-R | 10µM 2.5µL |
| water | 34µL | |
| polymerase | Phusion 0.5µL | |
| DNA | toothpick in glycerol stock of MG1655 | |
Transformations of Biobricks
As in the protocol given in the registry. Transformation of biobricks :
- BBa_B00300 pSB1A2 : RBS (well 3G plate 1)
- BBa_E0422 pSB1A2 : ECFP (RBS++Term) (well 11G plate 1)
- BBa_E0241 pSB1A2 : PoPs to GFP converter (well 15c plate 2)
- BBa_E0840 : (well 16E plate 1)
- BBa_J61047 pSB1A2 : Cre ORF (well 8P plate 4)
Transformation in DH5alpha subcloning efficiency Spread on LB-Amp