Vincent Parker Notebook
From 2007.igem.org
| Line 86: | Line 86: | ||
[[User:Vparker|Vparker]] 18:22, 12 July 2007 (EDT) | [[User:Vparker|Vparker]] 18:22, 12 July 2007 (EDT) | ||
Richard gave me my sequences and compared them. I had success with the T7-RBS. 2 out of 4 samples had what I needed but I did not number my samples of I need to send them off again just to verify which ones are which. My otsB-RBS came back perfect. My otsA-TT sequences actually came back coding for otsB-RBS so I either gave them all otsB-RBA and no otsA-TT or I actually have no otsA-TT and just otsB-RBS and I need to make some otsA-TT. I then took my plates from the RA and LB and colony PCRed that. The picture I took of it after was dismal. I was expecting 2k bands which in reality I somehow got a 1k band in each well. Meaning I have all parent vectors or.... its only otsB-RBS which would be closest with that band. Went to pizza lunch provided by Kate and after I transformed the otsBA in 1144 into lefty competent cells to only check if the plasmid is O.K. The sequences from otsBA the PCR came back with the same point mutation!!! This has been very troublesome since my other otsB that I originally PCRed did not have this mutation. I have traced the roots to template provided to me by Chris. I had an original tube which gave me my separate otsA and otsB. I ran out of the tube before I could use it for otsBA and Chris supplied me with a new tube from which I have yet to have a mutation free otsB come out of. I then decided to PCR from my original otsB and I actually had some left mover V5-7 in my box which I just then continued to sew to 6-4 which by my calculations should not have the point mutation I loath so much. | Richard gave me my sequences and compared them. I had success with the T7-RBS. 2 out of 4 samples had what I needed but I did not number my samples of I need to send them off again just to verify which ones are which. My otsB-RBS came back perfect. My otsA-TT sequences actually came back coding for otsB-RBS so I either gave them all otsB-RBA and no otsA-TT or I actually have no otsA-TT and just otsB-RBS and I need to make some otsA-TT. I then took my plates from the RA and LB and colony PCRed that. The picture I took of it after was dismal. I was expecting 2k bands which in reality I somehow got a 1k band in each well. Meaning I have all parent vectors or.... its only otsB-RBS which would be closest with that band. Went to pizza lunch provided by Kate and after I transformed the otsBA in 1144 into lefty competent cells to only check if the plasmid is O.K. The sequences from otsBA the PCR came back with the same point mutation!!! This has been very troublesome since my other otsB that I originally PCRed did not have this mutation. I have traced the roots to template provided to me by Chris. I had an original tube which gave me my separate otsA and otsB. I ran out of the tube before I could use it for otsBA and Chris supplied me with a new tube from which I have yet to have a mutation free otsB come out of. I then decided to PCR from my original otsB and I actually had some left mover V5-7 in my box which I just then continued to sew to 6-4 which by my calculations should not have the point mutation I loath so much. | ||
| + | |||
| + | |||
| + | [[User:Vparker|Vparker]] 14:52, 19 July 2007 (EDT) | ||
| + | [[User:Vparker|Vparker]] 14:52, 19 July 2007 (EDT) | ||
| + | [[User:Vparker|Vparker]] 14:52, 19 July 2007 (EDT) | ||
| + | [[User:Vparker|Vparker]] 14:52, 19 July 2007 (EDT) | ||
| + | [[User:Vparker|Vparker]] 14:52, 19 July 2007 (EDT) | ||
| + | [[User:Vparker|Vparker]] 14:52, 19 July 2007 (EDT) | ||
Revision as of 18:52, 19 July 2007
My Construction Files
My Sequencing Files
Vparker 17:11, 20 June 2007 (EDT) The sequences of the oligos I designed are as follows:
VP1AF-cgtaaAGATCTATGAGTCGTTTAGTCGTAGTATC VP2AIF-CGATGACATTATCTGGATtCACGATTATCACCTG VP3AIR-CAGGTGATAATCGTGaATCCAGATAATGTCATCG VP4AR-cgtatCTCGAGttaGGATCCTTACGCAAGCTTTGGAAAGGTAGC VP5BF-cgtaaAGATCTATGACAGAACCGTTAACCGA VP6BIF-GCAACGTATTACTCAGATtTGGCCACAAATGGCG VP7BIR-CGCCATTTGTGGCCAaATCTGAGTAATACGTTGC VP8BR-cgtatCTCGAGttaGGATCCTTAGATACTACGACTAAACG
The oligos were named according to my initials, numerical sequence, and letters determining direction of oligo. Ex. VP1AF- Vincent Parker 1 otsA Forward.
Vparker 15:04, 21 June 2007 (EDT)
After a long wait the oligos that I have ordered have arrived. I ordered 8 which PCR the two genes and get rid of the internal restriction sites. The template used to make the oligos was called MG 1655. In the ApE file which was split into two genes they were called ostA and otsB. They came in powder form and we have to dilute them down to 10 micro mols. That makes my master supply of my oligos when i use them i have to dilute them down to 100 micro mols. Using my oligos I can determine my PCR products... otsA will be broken up into two parts; PartA is 413 bps and PartB is 1077 bps. OtsB; PartA 485bps, PartB 381bps. I will PCR them overnight and digest tomorrow. Part A of otsA will be called V1-3 because i am using oligos 1 and 3 to PCr that specific part. Part B of otsA will be called V2-4. Part A of otsB will be called V5-7 and part B of otsB will be called V6-8.
VParker 16:37, 22 June 2007 (EDT) Big day planned. I have to take out my tubes V1-3, V2-4, V5-7, and V6-8 from the PCR machine.These are the 4 parts to my two genes. V1-3 is Part A of my otsA gene, it ends at the internal restriction BamHI site. V2-4 is part B to my otsA gene and it starts with the BamHI restriction site. I have already designed my oligos which code for a silent mutation within the BamHI site and codes for Leucine. V5-7 is part A of my otsB gene and ends at an internal BglII restriction site. V6-8 is part B of my otsB gene and starts at the BaglII restriction site and I have designed the oligos for that gene to have a silent mutation in the BglII site that codes for Isoleucine. As i came in to PCR my two genes, otsA and otsB, with the 8 oligos i designed. Two oligos per part. The oligos are named by number so when I mention V1-3 i am referring to part A of otsA but it will be PCRed with the first and 3rd oligos. Unfortunately when i came into the lab it was flooded and all I managed to do was take out Hannah's tube and my four out of the PCR machine and put them into the freezer for Monday. I did manage however to go home and sleep some more :-)
Vparker 16:02, 25 June 2007 (EDT)
Today i came into the lab and found that 3 out of the 4 PCR tubes were in the freezer but that i have lost the tube marked V6-8. I had the PCR products from the tubes V1-3, V2-4, and V5-7. After having fumed a bit over my lost PCR I went to work on setting up a new PCR tube for V6-8 and to also clean tubes V1-3, V2-4 and V5-7. Austin was kind enough to share the PCR machine with me and i put in the tube V6-8 in and also the sewing reaction i was working on named V1-4. V1-4 is a combination between V1-3 and V2-4. Since V1-3 and V2-4 are two parts of one gene and I had to PCR out the internal restriction site.Decided to try my hand in Agar plates today so I volunteered the High School team to learn how to prepare and pour them since Arthur was not here. They turned out looking good with no bubbles. So i was behind today in terms of my PCR and i have to digest and ligate tomorrow instead of today. Losing the V6-8 tube set me back an hour and a half so i will save the rest for tomorrow.
Vparker 15:04, 26 June 2007 (EDT)
Started off today with digestion of tubes V1-4 and V5-8 which are my genes that i PCRed. V1-4 was my otsA gene and V5-8 was my otsB gene. Atfer storing them in cultivation for an hour and a half Sam showed us how to prepare Agar solution and broth. we just put them in the autoclave machine and are waiting for both digestion and the autoclave to finish. There were problems with the autoclave because maybe the person who used it before us did not reset it after. Our Agar jars had to sit out while the machine exhausted itself and reset to take in our bottles. Nhu and I poured over 5 sleeves to help out the lab. The digestion of my tubes went well there were no problems and afterward I had to gel extract the DNA. I did not take a picture because too much exposure to UV rays is dangerous to my DNA and I wanted to keep as much of it as I could for the ligation process after I cut out the bright DNA bands from the gels. I had to melt the gels and put them through gel extraction recovery kit which binds the DNA to a filter and washes out all the enzymes and other solutions not wanted when I ligate the DNA to the vector. The number on the vector tube was 9145 and I named my ligation tubes otsA9145 and otsB1945. After ligation I had to heat shock the competant E.coli cells so that their membrane would become passive to the plasmid which contained my genes. I then used the little glass balls and smeared my competant cells with now hopefully my plasmid inside them. Tomorrow after I come in i will see if my bacteria grow. If not i am a complete failure :-(
Vparker 14:57, 27 June 2007 (EDT)
Vparker 16:37, 28 June 2007 (EDT)
Vparker 15:20, 29 June 2007 (EDT)
Sequences came in today with complete success in terms of my inserts being inside the plassmids and the cloning process was perfect. Today I have to design my operons using 6 basic Biobrick parks. My operon will look like this:
T7(promoter)-AGGAGG(Rbs site)-otsB-AGGAGG(Rbs site)-OtsA-terminator site.
The T7 promoter will make it transcribe a lot since we want the bacteria to produce a lot of the Trehalose sugar which will allow it to recover form freeze drying. We want otsB to be expressed more thats why its in front and then HOPEFULLY our Rbs sites will work. If not I will have to go to the library to experiment with others. I also clone saved my otsA and otsB. I have transformed everything I need for the operon into plates and they should be ready on Monday. I was also given an extension on my project. Besides the operon i designed Chris also wants me to form the operon of how it naturally exists in the E.coli cell. In the E.coli there is no Rbs site inbetween the otsB and otsA and they actually overlap a little which means I will have to sew together a 3 piece gene. The operon looks like this.
T7(promoter)-AGGAGG(Rbs site)-otsBotsA-terminator site.
Vparker 12:45, 2 July 2007 (EDT)
Vparker 13:34, 3 July 2007 (EDT)
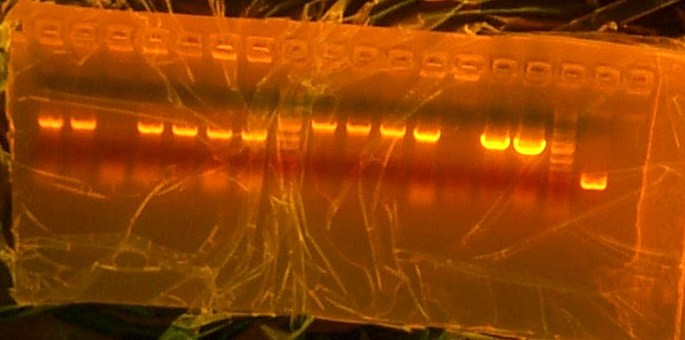
I took out the liquid media and mini-prepped my 5 samples. I then gelled them all to see if my transforming worked properly. After gelling I went to gel extract each one and they all had bright bands. The only problem was that otsA only had one band and it was supposed to have two. So I cut each one out and gel purified anyways but i set up an overnight digestion of otsA so that in the mornign I could re-gel it and re-gel extract it. As for otsBA I digested, then extracted and then ligated into 9145 plasmid and transformed into competent cells. I should have colonies for tomorrow to put into liquid media and then later to prep.
Vparker 13:34, 4 July 2007 (EDT)
Came in rather early just to get work done fast. I helped out Nhu by mini-prepping some of her liquid medias for her while i was gel purifying otsA and waiting to put otsBA into liquid media. OtsA did in fact have 2 bands this time around so I extracted from the gel, purified it and then brought out the T7, RBS,otsB, and TT purified end results from PCR clean up. I then transformed T7, otsA, and otsB into lefty competent cells and transformed RBS and TT into righty competent cells. Took out a colonized plate of otsBA and went straight into liquid media.
Vparker 13:34, 5 July 2007 (EDT)
Vparker 13:34, 6 July 2007 (EDT)
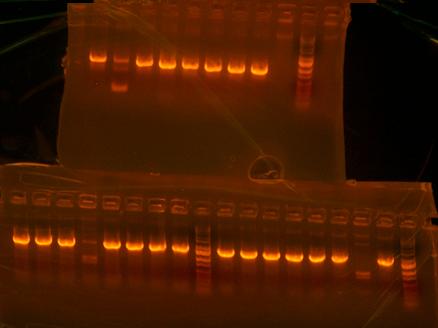
A taste of failure again. Could not gel the digest of the T7 to analyze to see if bands matched. All I managed to see BOTH times were just nice black bands and no DNA whatsoever. Then I colony PCRed the composite parts of LT(lefty T7), LB(lefty otsB), and RA(righty otsA) and the bands did not match up with the bands I wanted. I needed to start the cloning process over again. I transformed some more T7 because I used most of it trying to see bands. Need to put into liquid media and mini-prep to have new stock of T7. I did another digestion of T7 and used a different buffer and used more insert and did actually get a gel which showed that the T7 was a good plasmid. Also made my construction file of otsBA. Left an overnight digestion of my new composite parts which I will ligate and transform on Monday.
Vparker 15:10, 9 July 2007 (EDT)
Needed more otsA and otsB in the lefty for my composite parts so i miniprepped more from my stock. I ligated the overnight digestion of the composite parts and put them into lefty and righty E.coli competent cells. Needed to gel extract before ligation. Also need to send off otsBA my original PCR for sequencing because I got back the sequences for my forward and reverse primers and I unfortunately had a point mutation that was not silent. And I need to send it off for sequencing to see if it is from my original PCR product. Also need to put T7 into liquid media to mini prep tomorrow for Tulga.
Vparker 17:50, 10 July 2007 (EDT)
Mini-prepped more T7 105 wild type for Tulga and gave him a new stock and started to mini-prep otsBA but then sequences came back with the same point mutation and so T just threw away the otsBA tubes. I then re-PCRed V5-7 from my original template Mg1655 to try to cleanse the point mutation out. After PCRing V5-7 again I gelled it along with the conolyPCR product of otsB-RBS and otsA-TT. I also had T7-RBS but I needed to make a special gel with twice the amount of agarose in it to have the thickness needed so that the small band wouln't travel too fast. I then waited until before I left to transfer X's I made of the conolyPCR's bacterial colonies into liquid media so that tomorrow I would have usable mini-prepped stocks of my composite parts for tomorrow.
Vparker 18:22, 11 July 2007 (EDT)
Gel extraction right when I got to work. Extracted the PCR cleanup of 5-4 which was the finished product of 5-7 and 6-4. I then sent it off for PCR sequencing to see if I managed to defeat that pesky point mutation. Needed to digest lefty otsB-RBS and righty otsA-TT in order to be able to ligate them together. But then I found out we were out of plasmid so I digested some plasmid in BglII and XhoI so that I could refill the stock for the lab. I digested my lefty otsB-RBS with EcoRI and BamHI. I digested my righty with EcoRI and BglII. My left will turn into my insert and my righty will turn into my vector. After digestion gel extraction was necessary to purify out the bands I needed. The lefty I took the smaller band and the righty I took the only band because the other band is too small to see. Problem came with the bands form my digestion of the Bca1144 plasmid which should have had two bands but instead had 3 bands with two of them very close together. So I took the band that was closer to 2k which was the one that was originally supposed to be taken. Digested otsBA too with BglII and XhoI so that it would be able to be ligated with Bca1144. Cut out that band with no problems. Ligated together LB(lefty otsB-RBS) and RA(righty otsA-TT). After that ligation sat for a half an hour I heat killed the ligase and then digested again with BglII and BamHI to take advantage of the methylated sites from the lefty and righty competent cells. I then transformed the digestion straight into righty competent cells to grow for the next morning.
Vparker 18:22, 12 July 2007 (EDT)
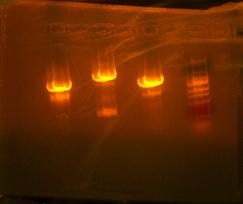
Richard gave me my sequences and compared them. I had success with the T7-RBS. 2 out of 4 samples had what I needed but I did not number my samples of I need to send them off again just to verify which ones are which. My otsB-RBS came back perfect. My otsA-TT sequences actually came back coding for otsB-RBS so I either gave them all otsB-RBA and no otsA-TT or I actually have no otsA-TT and just otsB-RBS and I need to make some otsA-TT. I then took my plates from the RA and LB and colony PCRed that. The picture I took of it after was dismal. I was expecting 2k bands which in reality I somehow got a 1k band in each well. Meaning I have all parent vectors or.... its only otsB-RBS which would be closest with that band. Went to pizza lunch provided by Kate and after I transformed the otsBA in 1144 into lefty competent cells to only check if the plasmid is O.K. The sequences from otsBA the PCR came back with the same point mutation!!! This has been very troublesome since my other otsB that I originally PCRed did not have this mutation. I have traced the roots to template provided to me by Chris. I had an original tube which gave me my separate otsA and otsB. I ran out of the tube before I could use it for otsBA and Chris supplied me with a new tube from which I have yet to have a mutation free otsB come out of. I then decided to PCR from my original otsB and I actually had some left mover V5-7 in my box which I just then continued to sew to 6-4 which by my calculations should not have the point mutation I loath so much.
Vparker 14:52, 19 July 2007 (EDT)
Vparker 14:52, 19 July 2007 (EDT)
Vparker 14:52, 19 July 2007 (EDT)
Vparker 14:52, 19 July 2007 (EDT)
Vparker 14:52, 19 July 2007 (EDT)
Vparker 14:52, 19 July 2007 (EDT)