Austin Day Notebook
From 2007.igem.org
| Line 24: | Line 24: | ||
* Pick colonies from single clone transformations. | * Pick colonies from single clone transformations. | ||
* Pick colonies from the hem gene + terminator transformation. | * Pick colonies from the hem gene + terminator transformation. | ||
| + | |||
| + | |||
| + | |||
| + | * Thoughts: | ||
| + | ** The hemoglobin expression wasn't significant when I tested it with the biobricked hemoglobin construct. All of the pellets were very dark, including the negative control. I'm not sure how to interpret that, but it isn't what I wanted to see. I'll still keep them around to run in the protein gel. | ||
| + | ** So the petDUET construct seems to work, but for what reason? The scar sites? A specific part? RBS? Promoter? | ||
==[[User:AustinDay|AustinDay]] 14:25, 3 August 2007 (EDT)== | ==[[User:AustinDay|AustinDay]] 14:25, 3 August 2007 (EDT)== | ||
Revision as of 09:03, 4 August 2007
My Construction Files
My Sequencing Files
Alcohol Detox Side Adventure
Other Detox Side Adventure
AustinDay 04:44, 4 August 2007 (EDT)
- Morning: (~2-2.5 hrs)
- Gel purify, ligate, and transform the single clones from the digestion.
- Gel purify, ligate and transform the hem genes into the terminator plasmid.
- Pick colonies of I716095-8.
- Take 1092 culture out to mini prep later.
- PCR the hemoglobin cassette for insertion into the petDUET plasmid.
- Digest petDUET for hemoglobin cassette insertion and alcohol dehydrogenase insertion.
- Digest pcr from ahdE. (run test gel beforehand)
Afternoon: (~30-45 minutes)
- Digest hemoglobin cassettes for insertion into petDUET.
- Pick colonies from single clone transformations.
- Pick colonies from the hem gene + terminator transformation.
- Thoughts:
- The hemoglobin expression wasn't significant when I tested it with the biobricked hemoglobin construct. All of the pellets were very dark, including the negative control. I'm not sure how to interpret that, but it isn't what I wanted to see. I'll still keep them around to run in the protein gel.
- So the petDUET construct seems to work, but for what reason? The scar sites? A specific part? RBS? Promoter?
AustinDay 14:25, 3 August 2007 (EDT)
- The analytical gel on the I716088 clones said that the #2 clone is good, so I took that to the next step to create the petDUET construct for best yield. (I716092)
- I digested the cytochrome cassette for insertion into the 4 mutant hemoglobin cassettes in biobricks format. I ligated and transformed those. For the I716083 (unmodified hemoglobin) I didn't have a pool, so I digested a single clone of that, for the I716084/5/6, I had pools, so I went ahead and used them for the construction. I figure the one single clone is a backup in case the pools fail horribly so that I can still test the cytochromes with the hem genes for monday. Now that I think about it though, I'm going to follow up with single clones of I716084/5/6, I'll digest those overnight, along with another aliquot of the cytochrome cassette.
- As of now, the western is planned for monday/tuesday and will include the biobricks as well as the petDUET of the hemoglobin expressing cassettes with supplemented hemin, as well as the biobricks construct with the hem and cytochrome cassettes.
- I'm redoing all of these because this was the gel I got today...
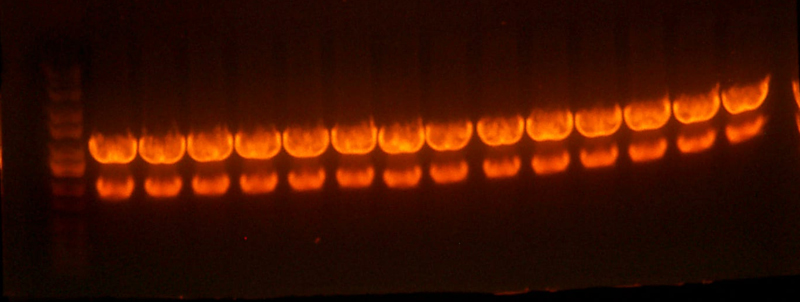
- Ran a gel of the one 92, 99, and 100 (I716XXX) parts.
- Lane 1: Ladder
- Lane 2: I716092 2-1
- Lane 3: I716099 1-1
- Lane 4: I716099 1-2
- Lane 5: I716099 1-3
- Lane 6: I71609100 1-1
- Lane 7: I71609100 1-2
- Lane 8: I71609100 1-3
- Lane 9: I71609100 1-4
- Lane 10: I71609100 2-1
- Lane 11: I71609100 2-2
- Lane 12: I71609100 2-3
- Lane 13: Ladder
- ALL WRONG SUCKA!!
AustinDay 14:50, 2 August 2007 (EDT)
- Didn't update yesterday, was kinda busy. Here's the report for yesterday:
- I grew up in equal volumes and equal starting amounts of the I716091/93/94, induced and added hemin at ~mid log and grew for 3 more hours then harvested. I pelleted them and the control looked significantly lighter than the others. I suppose that's the first sign of goodness. The iptg concentration was ~.5mM and the hemin concentration was ~.34mM.
- I also took the construction of the heme+cytochrome+hemoglobin cassette to the next step.
- Today:
- I had a very low yield of colonies today, but at least 1 of each construct.
- I made a liter of destain by 100ml methanol, 100ml acetic acid, 800ml DH20. (From: http://www.fhcrc.org/science/labs/hahn/methods/biochem_meth/comassie1.html)
- Also made some more loading buffer for the protein gels by adding, (the ratio, I made 3.5X of this) 2.4ml 1M tris, 3ml 20% SDS, 3ml 100% glycerol, 1.6ml beta-mercaptoethanol (smelly!!!!). (From: http://www.ciwemb.edu/labs/koshland/Protocols/Protein/sdspage.html
AustinDay 23:24, 30 July 2007 (EDT)
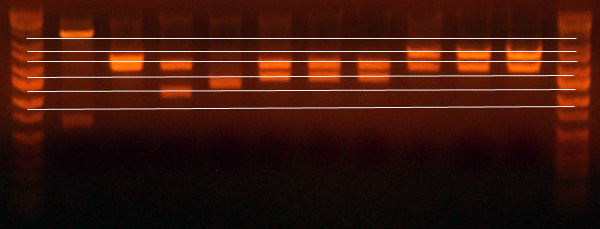
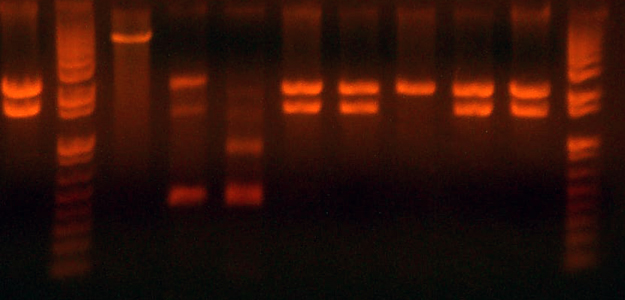
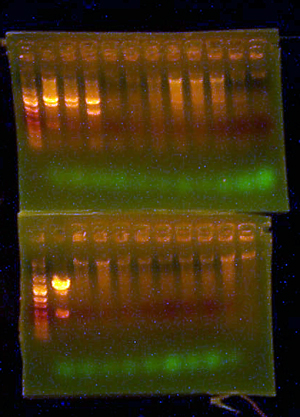
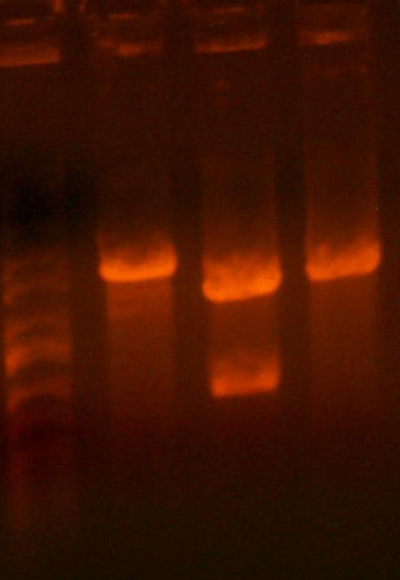
- Here is the gel from the clones I picked from what should be the complete petDUET cassette. Here are the lanes and results.
- Lane 1: Ladder
- Lane 2: I716091 #1 Expected ~500, Correct
- Lane 3: I716091 #2 Expected ~500, Correct
- Lane 4: I716091 #3 Expected ~500, Correct
- Lane 5: I716091 #4 Expected ~500, Correct
- Lane 6: I716093 #1-1 Expected ~1000, Wrong
- Lane 7: I716093 #1-2 Expected ~1000, Wrong
- Lane 8: I716093 #2-1 Expected ~1000, Correct
- Lane 9: I716093 #2-2 Expected ~1000, Correct
- Lane 10: I716094 #1 Expected ~500, Wrong
- Lane 11: I716094 #2 Expected ~500, Wrong
- Lane 12: I716094 #3 Expected ~500, Correct
- Lane 13: I716094 #4 Expected ~500, Correct
- Lane 14: Ladder
- So I took the I716091 #1, I716083 #2-1, and I716094 #3 clones, sequenced them as best I could (because one has a double alpha subunit) and I transformed them into petDUET.
- Also took kristin's hemA/B construct and I'm going to add that into my cassette. I'm hypothesizing that the heme biosynthesis bottleneck is significant and could be contributing to the lack of color. However...now that I think about it, the antibodies should still bind to the misfolded hemoglobin proteins even if there weren't enough heme. mmm...
AustinDay 19:17, 29 July 2007 (EDT)
- I grew up Madhvi's colonies, the DH10B's in LB and the others in Cam.
- I grew up 4 of each type of clone of I716091/3/4 (petDUET cassette)
- I've miniprepped the colonies I grew up yesterday of the petDUET beta insertions. I'm hoping to digest them and ligate and transform them today. I'll send things for sequencing tomorrow.
- I've tried a few programs that look at mRNA secondary structure. But I'm not sure how to identify termination hairpins or detremental secondary structures. For transcription termination, I imagine I can look at various types of terminators, some of which must work by hairpin formation. However, I'm not sure how to identify what kind of secondary structure would stop translation.
- Update from previous entries: I only got one more good colony that is a I716090 type. I'm redoing the pcr digestion for all of them, in case the ones that seemed good were bad. I think I may have forgot to add DpnI when I digested it the first time, which explains the very low yield.
AustinDay 19:07, 28 July 2007 (EDT)
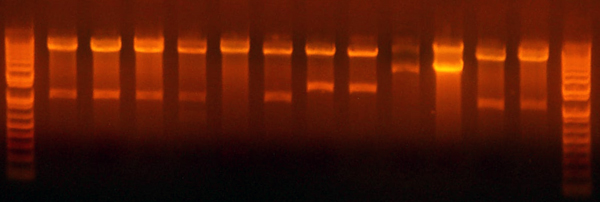
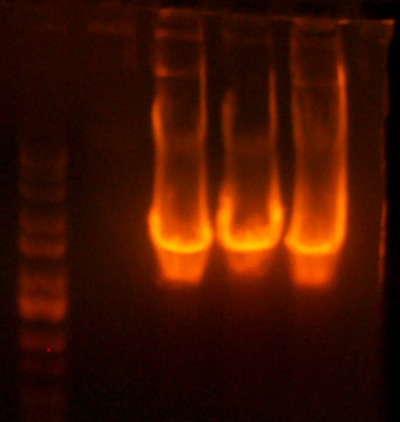
- I did the digest for the petDUET beta insertion plasmids. Turns out that the yield is pretty low. Here's the analytical gel.
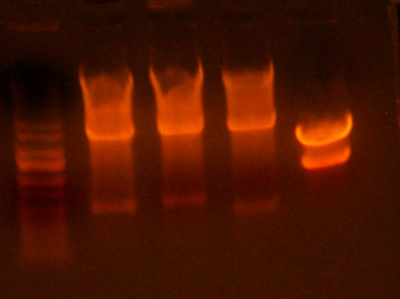
Lane 1: Ladder
Lane 2: I716087 #1 Correct
Lane 3: I716087 #2 Wrong
Lane 4: I716088 #1 Wrong
Lane 5: I716088 #2 Wrong
Lane 6: I716089 #1 Correct
Lane 7: I716089 #2 Correct
Lane 8: I716090 #1 Correct
Lane 9: I716090 #2 Wrong
Lane 10: Ladder
- This analytical gel digested the portions around the beta subunit, so there should be a band at ~500, which there is for I716087 #1, I716089 #1 and #2, and Possibly for I716090 #1. (Gotta look closely, very faint) I digested those with NdeI/BglII and cut them out. I ligated them with the corresponding alpha subunit already digested previously.
- I grew up more colonies of the parts that didn't have a band at ~500. I'll mini those and do another gel on them tomorrow, basically trail behind the good parts today by one day. I'll sequence a bunch of these guys on monday becuase quintara doesn't work on weekends.
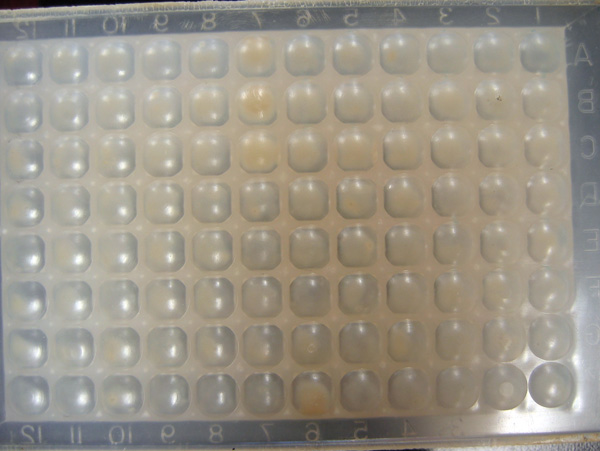
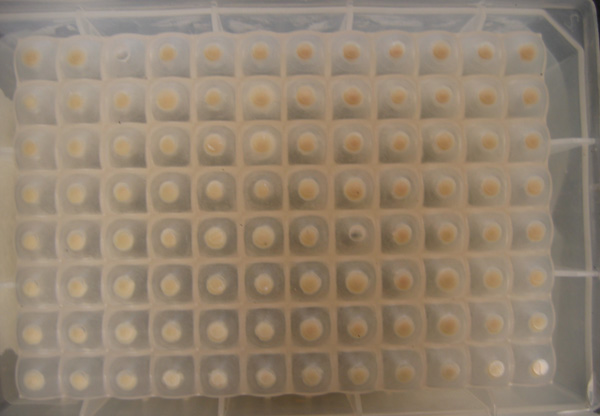
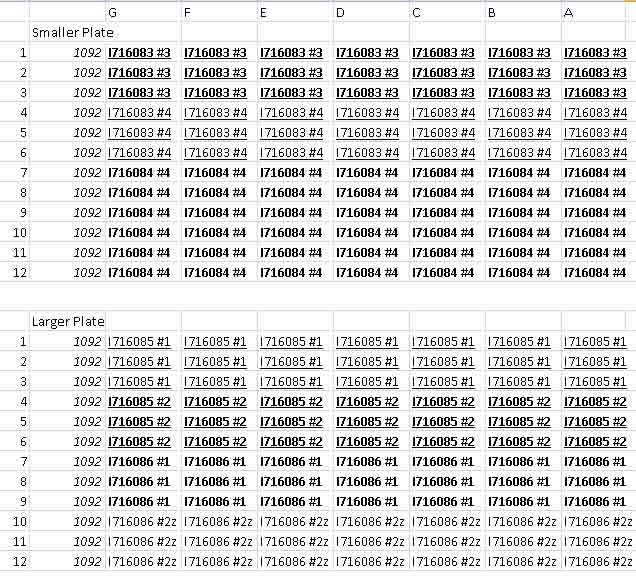
- And here are the results of the 96 well assay. (Not good...)
- Smaller plate (Control row is the top one)
- Larger plate (Control row is the bottom one)
- The 96 well plate showed no colonies that looked significantly different from the controls. Not the best outcome...
- So these were transformed from a sequenced plasmid that was confirmed to have the correct cassette to express hemoglobin into BLR strain and plated. The plate appeared to have a good number of colonies. These colonies were picked and used in the 96 well plate assay. Possibilities for incorrectness?
- Secondary structure is forming somewhere in the cassette and is prevent its transcription/translation.
- To test a possible secondary structure complication due to the biobricks format, the next attempt if this is the case is to put the parts into the petDUET plasmid and test expression there. If there is significantly more expression in petDUET, then that would indicate that it's a problem with the biobricks format or the parts other than the hemoglobin subunits that I am using. If there is still a problem in the petDUET plasmid, it may be because of a secondary structure consideration within in the subunits themselves. I'll see if I can find any online programs that might be able to test this hypothesis out in the meantime.
- Secondary structure is forming somewhere in the cassette and is prevent its transcription/translation.
- Hemoglobin is being expressed, but at levels too low to detect by eye in a pellet, possibly due to a bottleneck in the heme biosynthesis pathway or possible toxicity issues.
- To test this possibility, I should include a strain with and without the heme genes from Kristin on a protein gel. This is the only way to distinguish the redness being from heme, from actual hemoglobin. I'm surprised if this were the problem because I did add extra heme and iron to the induction solution I used for the 96 well assay, so I would bet against this possibility. As for the gel, I'll pick some more colonies from the plate which I used for the 96 well assay. (I'll also do an analyt ical digest in the meantime to make sure that they have the right sized cassettes, which would be a strong indicator that they have the correct cassette.)
- I don't think that toxicity is the problem here. I've previously grown up an induced colony in a previous 96 well plate, plated that, then grew that up in media, and the cassette was still perfect. So I don't think the cells are dying.
- Hemoglobin is being expressed, but at levels too low to detect by eye in a pellet, possibly due to a bottleneck in the heme biosynthesis pathway or possible toxicity issues.
- My best guess is that there is something wrong with the secondary structure, but that seems like such a bad excuse to me, not sure why, but I don't like it. I'll see if I can verify that secondary structure prediction using some programs online though.
AustinDay 12:10, 27 July 2007 (EDT)
- I grew up 2 96 well plates of the various mutants that should have the hemoglobin cassette in BLR. Induced after 3 hours and left in the 37.
- I also grew up 3 of each of the beta insertions of the petDUET plasmid. I miniprepped them and digested them overnight.
- Induction solution for this round:
160 ul IPTG 1M
100ul of iron
7ml lb
3.75ml hemin (filtered)
Add ~50 ul to each (About .75 mM)
AustinDay 14:02, 26 July 2007 (EDT)
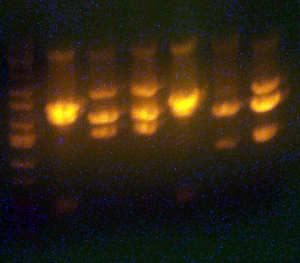
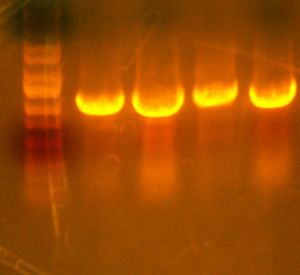
- Lane 1: I716085 cpcr #6 Good
- Lane 2: Ladder
- Lane 3: petDUET Good
- Lane 4: I716058 Good
- Lane 5: I716084 Good enough
- Lane 6: I716085 cpcr #1 Good
- Lane 7: I716085 cpcr #2 Good
- Lane 8: I716085 cpcr #3 Bad
- Lane 9: I716085 cpcr #4 Good
- Lane 10: I716085 cpcr #5 Good
- Lane 11: Ladder
- So it appears as though I can't do a colony pcr. Great. In any case, there are multiple good hits and I'll choose 2 of them to sequence.
- I cut out the correct sized band for the petDUET, I716085, and I716084 plasmids. I'll take those on to the next step.
- I also transformed the hemoglobin cassettes into the BLR strains and plated them (instead of going straight to culture like last time, mistake!)
AustinDay 21:55, 25 July 2007 (EDT)
- I attempted 2 colony PCRs on a total of 15 clones picked from the I716085 plate to try to get another good plasmid to sequence. Something was wrong though because none of the colonies gave bands. I followed the protocol and double checked the program, but still nothing. For some reason I didn't really trust that, so I grew up a few colonies and miniprepped them. I didn't have enough time to run a gel, but I have the digestions for tomorrow morning ready.
- On the BLR hemoglobin cassette front, I grew up 4 cultures in the 37 shaker and 4 cultures in the 30 degree shaker until mid log, induced with a solution of iron, hemin, and IPTG, and I'm letting those go overnight. The clones are I716083, I716084, I716086, and 9145-1092 (a negative control). These are single clones with numbers associated with them. Hopefully this will show a phenotype. If not, I'll have to rely on putting them into the petDUET plasmid.
- That petDUET plasmid actually did grow yesterday, on the plate and in culture. I did a digest on that, but I didn't like how the digestion came out. Since the I716058 pcr (to add the restriction sites to insert the various parts into the petDUET plasmid) looked like it was the wrong part, I figure I must have switched something where. The bands weren't even the close or the right sizes. Since I'm redoing the pcr with the other I716058 clone, I"m also going to redo the petDUET digestion and the I716084 digest because I didn't like how they looked.
AustinDay 04:11, 24 July 2007 (EDT)
- The sequencing says that there is at least one good clone for each construct except for I716085, I'm going to do a colony pcr on 5 more clones from that plate and sequence the promising colonies.
- The oligos for the RBS library construction came in, but I suppose I should characterize a working plasmid before trying to tune it, so I'm going to delay that pcr for a little bit.
- I'm going to transform what appear to be the good plasmids into cultures and see if I can detect phenotypes under different growth conditions (Temperature, induction times, etc...) I'll use a plain BLR strain with 9145-1092 as the negative control.
- Got oligos for the petDUET insertion. However, the 1ul of plasmid that I transformed into culture...wasn't growing after 5 or 6 hours. I'm not sure what went wrong, I mean...it was just a transformation. I also did a bunch of other transofrmations into culture using the same media. The antibiotic is correct, as far as it is labeled. I did a little test to make sure it is the correct antibiotic.
AustinDay 15:48, 23 July 2007 (EDT)
- Did an analytical gel on those cultures I was playing around with. (previously induced, but brown for some reason), as well as the naive cultures in DH10B. Let's check this out:
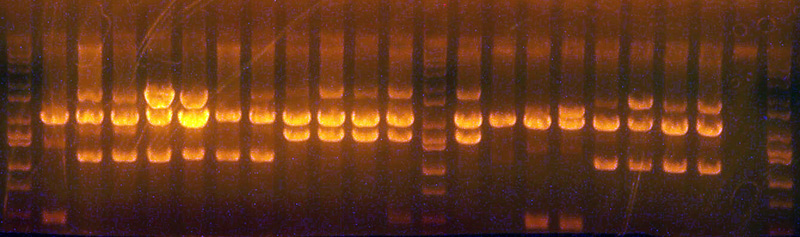
- The lanes and results are:
- Ladder
- I716084 #1z (Grown from the plate of the induced 96 well plate; grew brownish): Expected 2063/1522, Observed: Dark band ~2000 / light band ~1400 / light band ~500;
Conclusion: Wrong dude - I716086 #1z (Grown from the plate of the induced 96 well plate): Expected: 2063/1096, Observed: Medium dark band at ~3000 / Dark band at ~2000 / very light band at ~1750 / medium dark band at ~1100;
Conclusion: Partially cut, but mostly correct. Unexplained band at ~1750 - I716086 #2z (Grown from the plate of the induced 96 well plate; grew brownish): Expected: 2063/1096, Observed: Medium dark band at ~3000 / Dark band at ~2000 / very light band at ~1750 / medium dark band at ~1100;
Conclusion: Partially cut, but mostly correct. Unexplained band at ~1750 - I716083 #1 Naive: Expected: 2063/1096 Observed: Very dark band at ~2500 / Very dark band at ~2000 / medium dark band at ~1100;
Conclusion: Correct, but with an odd amount of uncut plasmid. - I716083 #2 Naive: Expected: 2063/1096 Observed: Very dark band at ~2500 / Very dark band at ~2000 / medium dark band at ~1100;
Conclusion: Correct, but with an odd amount of uncut plasmid. - I716083 #3 Naive: Expected: 2063/1096 Observed: Dark band at ~2100 / dark band at ~1100
Conclusion: Correct - I716083 #4 Naive: Expected: 2063/1096 Observed: Very light band at ~2900 / Dark band at ~2100 / dark band at ~1100
Conclusion: Correct, but with a slight amount of single cut plasmid - I716084 #1 Naive: Expected: 2063 / 1522 Observed: Very light band at ~3500 / Dark band at ~2000 / Dark band at ~1500
Conclusion: slightly partially cut, but looks correct - I716084 #2 Naive: Expected: 2063 / 1522 Observed: Medium dark band at ~3500 / Dark band at ~2000 / Dark band at ~1500
Conclusion: Significant is partially cut, but looks otherwise correct - I716084 #3 Naive: Expected: 2063 / 1522 Observed: light band at ~3500 / Dark band at ~2000 / Dark band at ~1500
Conclusion: Little partially cut, but looks correct - I716084 #4 Naive: Expected: 2063 / 1522 Observed: Medium dark band at ~3500 / Dark band at ~2000 / Dark band at ~1500
Conclusion: Some is partially cut, but looks correct - Ladder:
- I716085 #1 Naive: Expected: 2063 / 1522 Observed: Light band at ~3500 / Very dark band at ~2000 / Very dark band at ~ 1500
Conclusion: Some singly cut, but mostly correct - I716085 #2 Naive: Expected: 2063 / 1522 Observed: Very Dark band at ~2000 / Very light band at ~1300
Conclusion: Sadly, incorrect - I716085 #3 Naive: Expected: 2063 / 1522 Observed: Very dark band at ~2000 / Very light band at ~1400
Conclusion: Damn wrong - I716085 #4 Naive: Expected: 2063 / 1522 Observed: Very dark band at ~2500 / Very dark band at ~2000 / Very light band at ~ 1300
Conclusion: Frikin wrong dude - I716086 #1 Naive: Expected: 2063 / 1096 Observed: Very light band at ~3000 / Very dark band at ~2000 / Very dark band at ~1100
Conclusion: Slight singly cut, but mostly correct - I716086 #2 Naive: Expected: 2063 / 1096 Observed: Dark band at ~3000 / Dark band at ~2000 / Dark band at ~1100
Conclusion: Significant singly cut, but probably correct - I716086 #3 Naive: Expected: 2063 / 1096 Observed: light band at ~3000 / Very dark band at ~2000 / Very dark band at ~1100
Conclusion: Some singly cut, but mostly correct - I716086 #4 Naive: Expected: 2063 / 1096 Observed: Dark band at ~3000 / Very dark band at ~2000 / Very dark band at ~1100
Conclusion: Significant singly cut, but probably correct - Blank:
- Ladder:
- Overall conclusions: I716083 #3 and #4 look promising, I716084 #1 and #3 look promising, I716085 #1 looks promising, and I716086 #1 and #3 look promising. I'll also sequence that one that I (thought) I saw a significant phenotype with (I716086 #2z).
- That is all, good day.
AustinDay 20:19, 21 July 2007 (EDT)
- I also did a bit of price checking for accessory items for the jamboree thingie:
http://www.queensboro.com
(Logo embroidery only)
Polo shirts: 10-25 bucks
T shirts: 8-15 bucks
Hoodies: ~30 bucks
http://www.customink.com/cink/r.jsp?C=13&D=10200
Hoodies: ~31 bucks each
http://www.logosoftwear.com/hoodedsweatshirts.php
Hoodies: Many options, mid 30s and up. (Some look very nice)
http://www.uberprints.com/catalog/product/?id=GIG200
~16 t shirts 3 colors front only
~36 for hoodies, embroidered
http://www.logosoftwear.com/hoodedsweatshirts.php
hoodies ~35-38 bucks
http://www.shirtcity.com/shop/index.php?PHPSESSID=0e8f37d3f8d09127207dfed1c0157694&currblock=1&set_vorne_own_picture=True
~ 26 for hoodies
http://www.epromos.com/browse/Ne1-N11068-11603.html
~ 30 and up for hoodies
http://www.zazzle.com/custom/tshirts
t-shirts ~11 and up
ties: ~27 and up
http://www.screenprintit.com/
~15 for shirts
http://www.printfection.com/customer/custom.php?rootid=7&invoptid=111
shirts: ~11-15
http://www.superiorpromos.com/viewProduct.html?id=2651&ca_id=23_307_833_991&gclid=CI6yz4jYuY0CFQzDYgodgiRbLg
Shirts: ~15
http://www.logodogzprintz.com/orderdetails.asp
~11 bucks, 3 colors, front *
~21 bucks for hoodies, 3 colors, 7.5 oz *?
Pins:
http://www.ashlynpromotions.com/custom_lapel_pins_cloisonne.htm
Pricing: Cloisonne 100 200 300 500 750 1000 2000 3000 5000 10000
3/4" $2.57 $2.04 $1.49 $1.27 $1.09 $1.03 $0.95 $0.83 $0.71 $0.62
1" $2.60 $2.10 $1.60 $1.31 $1.14 $1.06 $1.02 $0.88 $0.80 $0.71
1.25" $2.71 $2.18 $1.68 $1.38 $1.22 $1.16 $1.08 $1.93 $0.83 $0.75
1.50" $2.94 $2.44 $1.93 $1.47 $1.32 $1.24 $1.18 $1.00 $0.86 $0.83
1.75" $3.13 $2.61 $2.12 $1.66 $1.49 $1.42 $1.36 $1.18 $1.04 $1.01
2" $3.35 $2.83 $2.34 $1.87 $1.70 $1.64 $1.56 $1.39 $1.25 $1.22
http://www.qualitylapelpins.com/coins.php
Call for quote
http://www.pinmart.com/request_menu.cfm
E-mail for quote
AustinDay 16:11, 21 July 2007 (EDT)
- Okay... so I think I've been slacking on the notebook a little these past few days because I came in today to 6 culture tubes that I don't remember growing up or where they came from. Fun. Two of them are reddish brown, but they're probably RFP. (update: they're probably just overgrown) I'm not sure how to tell the different between the two at this point. I compared regular cultures to the heme cultures that kristin have to the red tubes that I found, but they all look fluorescent under UV light. Eh...
- I retransformed what should be the complete hemoglobin cassettes for I716084/5/6. I'm going to pick a few colonies from each plate today and see if I can get an intact cassette.
- I also redid the ligation/transformation for the I716083 part, I got more colonies than the second try (this is the third), but still there were very few. Whatever, I'll grow some up and hopefully get an intact cassette. I'm not going to worry about the rbs diversity anymore since I can do that IE pcr method to introduce them into cassettes. (I don't know why I didn't think to do that earlier, it sounds much easier). I already ordered the oligos to do that. The only problem is again that there are two annealing points for the alpha subunit primers, but I included an annealing region on the other end of the oligo that will increase it's tendency to anneal to the correct region. I'll still get two bands, but like the other try with a similar oligo, the good sized band is cuttable.
- Oh wait, I remember where those mystery cultures were from. They were from the restreaks of promising colonies fro the 96 well plate. Mmmm....so some of them are brownish? RFP? Let's find out. (The 4:1 and 6:1 z clones were the darkest)
- Here's the analytical gel of the colonies I ran. 4:1 was brown and so was 6:2. (Digest was BglII/XhoI) The lanes are
- Marker:
- I716084 #1z
- I716084 #2z
- I716085 #1z
- I716085 #2z
- I716086 #1z
- I716086 #2z
- Kind of strange that lanes 3/4 would represent each other and 2/5 would also. Maybe I got a tube switched somewhere? In any case, there seem to be three different types of plasmids present. There is the one that has
a ~2000 and ~500 sized fragments. There is one that has ~2000 and ~1500 sized fragments. And there is one that has ~3000, ~2100, and ~1200 sized fragments.
- The ones with the ~500 sized fragments may be a single beta or alpha subunit plasmid. I'm not sure if a single subunit would give a phenotype... assuming that the promoter is intact. The ~1500 fragments seem to resemble a complete beta/di-alpha cassette. The ~1200 fragment in the last two lanes could be representative of the correct construct for the I716086 part, which would be mutant variants of the alpha and beta subunits. I feel that I should redigest these suckers before I send some for sequencing just because it looks like a lane got switched or something... I'll also retransform the I716086 #2 plasmid to see if I can get that dark phenotype again. (I'll also retransform the second to last lane and the I716083 #1 plasmid, in case I actually switch a tube somewhere)
AustinDay 15:22, 20 July 2007 (EDT)
- It's not a good day for hemoglobin.
- The clones I picked straight from the I716084/5/6 plates and sequenced came back either mixed or without the genes. I don't know who's fault that is, so I did an analytical digest with either BglII/XhoI or EcoRI/BamHI.
- Lane assignments:
- Ladder: (Starting from top left)
- I716084 library (The library transformed into the BLR strain)
- I716085 library (The library transformed into the BLR strain)
- I716085 library (The library transformed into the BLR strain)
- All the other non-ladder lanes are various clones of I716084/5/6 plasmids transformed into the BLR strain. (All of them look really crappy)
AustinDay 18:52, 19 July 2007 (EDT)
- So I did a 96 well plate of a bunch of the mutants that should be expressing hemoglobin. The sequencing verified that there is a promoter on them. The pellets were slightly discolored, but I'm not sure if that was due to the hemin I added or if it changed color on its own.
- I'm going to try growing up some of the darker clones in 5ml cultures without adding hemin to see if they are actually changing color. If they are in fact a different color, then I'm going to try to do a nother 96 well plate with only adding IPTG and then choosing the darker clones from that (without hemin). I'll isolate those clones and characterize them. I'll then use that to go ahead with adding the other parts and running the protein gels to quantify the hemoglobin.
AustinDay 00:47, 18 July 2007 (EDT)
- I put the I716084/85/86 plasmids into the BLR strains so we may see red tomorrow! S%!t! About time! (BTW: The I716071 transformation from yesterday worked like a charm.)
- I also am lagging behind one day trying to get the I716071 pool to the same stage as the other mutants.
AustinDay 19:14, 16 July 2007 (EDT)
- Grew up the I716071/74/75/76 pools. There were only 4 colonies on the I716071 plate, so I redid that ligation and transformation. I used 4.5ul of the fragments instead of 2, so hopefully that'll do it. I imagine that it's something else I did wrong becuase 4 is kind of a low number. I also used a 200ul of cells instead of 90. (update, after ~7 hours with TG1 cells, I can't really see any colonies. $*%&...) But at least the gel for the others looks good. I didn't want to take a picture or else I'd fry my DNA.
- As for the 74/75/76 parts, I got a decent number of colonies, so I'm taking those ahead and putting T7 promoters in front of them. (I716105). This is the wildtype and should be the strongest that we have.
- I also told the peoples making the accessory genes (AHSP, cytochrome stuffs, hem stuffs) To make complete cassettes so that I can include them without reduced expression. (Apparently having them all under the same operon reduces the expression of the genes near the end? Sure, why not.)
AustinDay 15:00, 14 July 2007 (EDT)
- I took the weekend off suckas!! But I got sequencing back and all of the beta portions are good. One had a silent point mutation, but that may have been there already and I just forgot to update the sequence file. I did that just now though. I took those beta portions along with the corresponding alpha pieces to form the I716071/74/75/76 composite parts. Once I can confirm that those libraries are good, I'll take that in many directions. One of which will be to attach a T7 promoter right away and test the expression with nothing else. I'll also see if I can get those other parts (AHSP, cytochrome b5 and b5 reductase, and the hemA/B/C/D genes in front of their own T7 promoters, but I'll be able to do that in parallel with putting a T7 in front of my own part.) Then I can take my part and add each of those other parts I just mentioned individually and see the effect on yield or color. Once I can get basic characterization and verify that the parts are in fact doing something, I'll start to add more until I have a complete cassette. Somewhere in there I'll have to run a protein gel with the hemoglobin antibodies. I'll wait until I get a good number of interesting phenotypes to fill up the 15 lanes of the gel.
- That is all, good day to you
- Oh yea, I also tried that new oligo I got to get the di-alpha amplification product. The band was significantly brighter, but there was still 2 there and they were pretty close. I tried my best, but I'm afraid there will still be significant background. Hopefully I'll still be able to get a good sequencing read.
AustinDay 14:44, 11 July 2007 (EDT)
- I think I figured out partially why some of the things failed. when doing the PCR on the di-alpha fusions, I forgot to take into account that the oligos for the alpha subunit will anneal to two different spots, creating less of the fragment I want (the di-alpha amplification product) and much more of the single alpha product. I ordered an oligos to solve this problem, but in the meantime, I'm going to try to cut out only the di-alpha band by doing a verrrry careful gel. We'll see how that works out.
- Okay, the gel seemed to cut out relatively clean, I also ligated and transformed the I716080, I716081, and I716082 parts. I'll paste them together with the alpha subunits tomorrow. But I'll make sure to take it slow and do a lot of colony pcrs and analytic gels. (No more rushing through things...)
AustinDay 22:03, 9 July 2007 (EDT)
- No red yet... DAMN!! I'm not sure what was wrong. I'm was going to sequence them, but there was some mini prepping complications...involving Mr. Yu. Anyway, I grew up those cultures again so I can mini prep them tomorrow and I also started the PCR that attaches the rbs library to the alpha and beta parts. (I'm starting over again, but I'm going to be much more careful.)
- When I say "starting over", I mean just from the first step of the libraries. I think I may have forgot to digest with DpnI during some PCRing. I doubt that was the main problem, but I've been through the construct and it seems sound. A lot depends on any sequencing results I get back, and/or the colony PCR I will run on them tomorrow if the sequencing doesn't give me anything useful.
AustinDay 17:27, 8 July 2007 (EDT)
- I got colonies for all of the plates, but there weren't very many for the I716084 or I716085 constructs. I suspect that it might be because the bands were very hard to see and I may not have a very high concentration of those two parts going into the ligation. I'm going to try it again with more of the insert.
- I grew up 5 clones of each of the cassettes which potentially can express hemoglobin. I'm gong to grow them up to about mid log at 37, then add IPTG, hemin, and FeCl3 and move them to the 30 degree room overnight. Hopefully they will be somewhat different color in the morning. (Hopefully a dark chocolate color)
- I made up a little "induction " solution, made from 125ul 1M IPTG, 100ul of ~50mM FeCl3, and 100ul of hemin. (Unknown concentration). This will make a concentration of IPTG that is ~.5mM when 4.5 of this solution is added to 5ml of culture.
- I also made another one with 125ul 1M IPTG, 25ul FeCl3, and 10ul of Hemin. (Because the other one was too dark, didn't like it.) I'll use this mixture for the first try.
AustinDay 04:25, 7 July 2007 (EDT)
- Okay, so yesterday I verified my sequencing and everything seemed to be alright. I completed the constructs for the three cassettes (beta / alpha) for the unmodified, highest yield, adn two best mutants.
- We're going to go in at 2am today and scrape and grow up colonies for tomorrow. Horray!
AustinDay 21:28, 5 July 2007 (EDT)
- I grew up the library pools of the I716080/81/82 parts and minied them and set up the PCR for the next step to construct the cassette with rbs-apha-rbs-beta, without scar sites and unmutated and 3 mutant varieties. These should give the unmodified hemoglobin, the highest yield mutant, and the two best candidates for actual use as a blood substitute.
- Quintara doesn't work on the weekends or on holidays, what lazy asses. I'm two steps into a construction and I don't even know if the first part is correct or not. I've been carrying along 3 clones of the providence beta mutant, so one of them better be correct of I'm going to be pissed.
- That is all, good day.
AustinDay 11:55, 4 July 2007 (EDT)
- Grew up I716077/8/9 colonies.
- Mini prepped the Providence single mutant betas for sequencing.
- Mini prepped the I716077/8/9 and sequenced them.
- Made parts I716080, 81, and 82.
AustinDay 12:56, 3 July 2007 (EDT)
- So I figured out another thing I was doing wrong... It involved making construction files in the past and then not using them to make the part, then going back to that construction file and trying it 4 times until I figured out it was wrong. Anyway, I fixed that, and this transformation to make the providence beta mutant should damn well work.
- I finally got my sequencing back from quintara, 3 out of 5 were correct. So I finally have both di-alpha fusions. All I need is this providence mutant to proceed with making all of the mutants for the protein gel. I hope some of those constructions turn red, or else I'll have to rethink the approach...
- The lanes I'm going to run in the next western are planned to be:
- (1)Ladder
- (2)Positive (+) control (Commercial hemoglobin, known concentration; yet to be determined)
- (3)Positive (+) control (Commercial hemoglobin, known concentration; yet to be determined)
- (4)Positive (+) control (Commercial hemoglobin, known concentration; yet to be determined)
- (5)Positive (+) control (My nose blood, to compare to the actual concentration of blood hemoglobin); Supernatent
- (6)Negative (-) control with 1092 (terminator) plasmid; Pellet
- (7)Negative (-) control with 1092 (terminator) plasmid; Supernatent
- (8)9145-I716071: pTrc-rbs lib-(NO SCAR)-beta-rbs lib-alpha (Unmodified everything); Pellet
- (9)9145-I716071: pTrc-rbs lib-(NO SCAR)-beta-rbs lib-alpha (Unmodified everything); Supernatent
- (10)9145-I716074: pTrc-rbs lib-(NO SCAR)-beta 1X mutant, providence (Lys82Asp) - rbs lib - unmutated di-alpha- (Best Yield); Pellet
- (11)9145-I716074: pTrc-rbs lib-(NO SCAR)-beta 1X mutant, providence (Lys82Asp) - rbs lib - unmutated di-alpha- (Best Yield); Supernatent
- (12)9145-I716075: pTrc-rbs lib-(NO SCAR)-beta 2X mutant, providence (Lys82Asp) and Presbyterian (Asn108Lys)-rbs lib - unmutated di alpha - (Best all around); Pellet
- (13)9145-I716075: pTrc-rbs lib-(NO SCAR)-beta 2X mutant, providence (Lys82Asp) and Presbyterian (Asn108Lys)-rbs lib - unmutated di alpha - (Best all around); Supernatent
- (14)9145-I716076: pTrc-rbs lib-(NO SCAR)-1X mutant beta, presbyterian (Asn108Lys)-rbs lib-2X mutant alpha (L29F and V96W) (Second best all around); Pellet
- (15) 9145-I716076: pTrc-rbs lib-(NO SCAR)-1X mutant beta, presbyterian (Asn108Lys)-rbs lib-2X mutant alpha (L29F and V96W) (Second best all around); Supernatent
AustinDay 12:45, 2 July 2007 (EDT)
- I plated the providence digest this morning, I'll pick colonies in about 8 hours.
- Current status on the alpha and beta for the new rbs construction:
Unmodified di-alpha: Have
2x di-alpha: Awaiting sequencing results
Have all betas except the providence 1x mutant.
- Goal: pTRC-No Scar-rbs-beta-rbs-alpha-TT:
Generalized dbbs strategy:
Alpha:
Digest 9145-bca1092 (Bgl2/EcoR1, 2206/3, Larger)
Digest (Alpha variant) (BamH1/EcoR1, ?/?, ?)
Product is Biobrick Part #.
PCR CA1150/CA1099R on Part #. (? Bp)
Digest (Bsa1/Bgl2) and ligate with beta portion.
Beta:
PCR CA1154F/G00101 on (Beta variant) (? Bp)
Digest (Bgl2/Xho1) and sub into pTRC (Bgl2/Xho1)
Product is Biobrick Part *.
PCR CA1150F/D58 on Part *.
Digest (Bsa1/BamH1) and ligate with alpha portion.
- There was a mismarking in the construction files regarding the oligos for the mutants... I fixed it, but it caused a lot of pain in the meantime.
AustinDay 12:28, 2 July 2007 (EDT)
- Yesterday I redid the quickchange to make the single mutant of beta "providence". I left the digestion overnight in the pcr machine because the last bus left at 8:05. (Damn bus!!)
- I also miniprepped and sequenced the potential 2X mutant di-alpha fusions. (Hurry quintara!!)
AustinDay 16:52, 30 June 2007 (EDT)
- Yesterday I basically redid the PCR for the ADtemp2 part, I also did a colony pcr to try to find some of the good 2X di-alpha fusions. There was one that was off by a point deletion in the sequencing, I think I can probably find a good one in there also.
- Today I redid the ADtemp1 step and digested it overnight (in the PCR machine).
- I grew up the likely candidates for the di-alpha fusion 2X mutant based on the colony PCR.
AustinDay 01:15, 28 June 2007 (EDT)
- Okay, I redid the ADtemp2 transformation from the first step. Hopefully it'll work this time.
- I re-transformed some mini preps that were important and/or were running low so that I can replace them. I don't really trust the clonesaver cards because of some of the things people on random message boards have said. I'll still use them, but I'll definitely have another backup.
- I mini prepped and sent for sequencing, the di-alpha fusion (mutant and unmodified), as well as the single beta providence mutation.
- I'll start making the correct mutants to see if I can get a good yield soon. These things better start turning red soon.
AustinDay 11:30, 26 June 2007 (EDT)
- I got very few colonies on both the ADtemp2 and the I716070 plate, it makes me think that the same thing was wrong with both. I grew up 4 of the colonies from the I716070 plate, but since the ADtemp2 plate is supposed to be a library, and I did a gel of the ADtemp1 pcr product, it makes me think that it might be the ptrc digestion I used, in addition to whatever else went wrong. In any case, I'm making a fresh ptrc digest and I'm going to reuse the pcr product, because I'm fairly certain that it's correct.
- I'm finishing up the quickchange to make the providence mutant, transforming that once the DpnI digestion is done.
- I'm also remaking the 092 and 053 digests, in case all 4 of the colonies I grew up were bad, which might be the case since I got so few colonies.
- Hemoglobin antibodies came in today!
- Planning for the best hemoglobin mutants: (More organized)
- Best yield mutant: Di-alpha globin / providence (beta-Lys82Asp)
- Best all around: Di-alpha / Providence (beta-Lys82Asp) / Presbyterian (Asn108Lys)
- Second best all around: Presbyterian (Asn108Lys) /alpha-L29F / alpha V96W
- Alpha subunit mutants:
Unmodified di-alpha fusion: I716068 (Need to make)
Alpha with L29F and V96W: I716062 (Have)
- Beta subunit mutants:
Providence (beta-Lys82Asp): I716072 (Getting today)
Providence (beta-Lys82Asp) + Presbyterian (Asn108Lys): I716063 (Have)
Presbyterian (Asn108Lys): I716059 (Have)
AustinDay 15:06, 25 June 2007 (EDT)
- I completed up to the plating of the ADtemp2 part for the making of the pTrc-rbs-beta part, which will be joined with the ADtemp5 part (-rbs-alpha-TT) later.
- I made the alpha-TT part today and transformed it into colonies.
- I started the second portion of the quickchange method for the creation of the providence mutation.
- Still waiting on the oligos for the di-alpha fusion creation.
- I figure at this point that I should make as many of the mutants and the "good" rbs library as I can so that when we get the HbA and antibodies, I can have some cultures that actually show a meaningful amount of hemoglobin. This would require that I also take the mutant alpha and beta basic parts and run a pcr on them with the oligos that chris made to avoid the scar site between the RBS and the part. Once I get the di-alpha fusion made, I'll do that with that. Once this quickchange is done, I can start that procedure for the mutants.
- So the goals for tomorrow:
finish the quickchange and plate that ASAP, use TG1s
Grow up the alpha-TT part and the ADtemp2 part.
Miniprep and sequence the two colonies I picked of the 2X mutants.
Wait for the beta mutant to make colonies, then grow some up overnight. Same for the alpha-TT
See if you can get a copy of the AHSP on a different antibiotic marker plasmid.
AustinDay 15:32, 24 June 2007 (EDT)
- Just planning out my method of attack for the next few days.
- Add description of mutants (providence etc...)
- "Providence(asp) mutation" (ßLys-82 -> Asp): Increases soluble output
- "Presbyterian mutation" (Asn-108 -> Lys) in beta-globin: Reduces oxygen affinity to a level comparable to erythrocytes. Decreases soluble output.
- Organize next set of experiments:
- To increase soluble output:
- Higher hemin concentration (final concentration used here was .63 for high end and .34 for low end) gave ~ 8% higher soluble protein.
- 28 degrees fermentation gave higher soluble protein than 30 degree fermentation. By ~6%.
- Di-alpha globin alone, compared with wild type beta and presbyterian , gave slightly higher soluble protein.
- Providence mutation increased soluble protein by ~9.5% with the presbyterian and 8% without.
- Best yield: Highest soluble protein configuration was di-alpha globin + providence mutation - presbyterian mutation.
- Best P50/yield combination: (Needs AHSP in order to have significant yield) Providence + Presbyterian + di-alpha linkage: P50 = 44. % Soluble = 17.5. % Total = 35.7
- Good Auto-oxidation, unknown yield, P50 = 34. Presbyterian + aL29f + aV96W
- Second tier attempts:
- b-N108D in place of presbyterian increase the P50 more than presbyterian. (For those with that mutation, try it out)
- Combination of surface modifications to increase P50 by 3x: Try using these in place of presbyterian.
- Val1 -> Met:
- His2 -> deleted
- Thr4 -> Ile
- Pro5 -> Ala
- Ala 76 -> Lys
(There are also many other mutations that will increase P50, yield is most important at this point)
- Try providence + di-alpha linkage + some surface mods.
- Get AHSP in there.
- Details of incubation and induction: (Baxter paper)
- On a pUC high copy number origin of replicaiton.
- Plasmid under a Ptac promoter.
- 15ug of tetracycline per ml
- Expression was induced by adding IPTG at concentrations between 10 and 200uM.
- Expression was induced during log phase. (OD 1/3 of final) (Final was ~30)
- Grown at 37 for 12-24 hours.
- Hemin was added every 3 hours after induction. (Although different concentrations didn't affect the cell densities.
- They added human serum albumin in 50mM Tris pH 7.5, such that the concentration did not exceed 50uM. (In HSA, the hemin forms a 1:1 complex that has a well defined absorbance spectrum at 625 nm. (Look into paper on baxter paper) They were able to calculate the amount of hemin being taken up over time.
AustinDay 16:48, 22 June 2007 (EDT)
- So we had the great flood today. Looks like a tornado went through the offices.
- Digested, ligated, and transformed the I716067 part (added the promoter onto the 2X mutant hemoglobins)
- Also ordered the oligos for the di-alpha hemoglobin 2X mutant fusion. I wonder when they would get in...?
- I pretty much can't do anything else until we get the hemoglobin antibodies and commercial grade hemoglobin, at least in lab. I know Chris has been trying to locate a machine we could use to measure the P50, I suppose I could read up on how that is done.
AustinDay 20:24, 21 June 2007 (EDT)
- Designed the oligos for the Di-alpha mutant fusion protein.
- Scraped and grew up the I716067 (-rbs-2X mutant alpha-rbs-2X mutant beta)
- Grew up 3 clones from the 20X dilution of 9145-1154 #1.
- Still waiting for the hemoglobin and antibodies... then the real fun can start.
AustinDay 12:45, 20 June 2007 (EDT)
- The sequencing came back and the first 1154 part was a dud, the second was a failed read. This is a bit annoying. I'm going to make a 1000X dilution of the #1 clone and retransform it. The read came back correct for that one, although the gel wouldn't work... But perhaps there's still a good one it there someplace.
- I also made mini preps of all of the chassis plasmids. (9104, 9106, 9023, and flu/inv)
- I also digested and transformed the I716067 part (The joined rbs lib-beta-rbs lib-alpha 2X mutant part)
- Did a protein gel of (ladder) (9145-1089 neg control) (I716061 #1) (I716061 #2) (I716061 #3) (I716061 #4) (I716061 #10) (I716061 #11) (I716061 #12) (I716061 #14) (9145-1089)
- It appears that there is a band at about 38KDa that is much darker on the I716061 clones and very light, but I believe detectable, on the 9145-1089 negatives. That paper that did a 15% SES gel got bands for hemoglobin at ~14 and 13 KDa. There are bands that are present on all of the lanes, however, there are two which are significantly darker than any of the others, and both of those are the I716061 clones. These are in the correct position (13-14 KDa). Maybe they are different levels of hemoglobin? Chris suggests that we run a higher percentage acrylamide gel next time to be able to resolve such small bands.
AustinDay 12:20, 19 June 2007 (EDT)
- Grew up some of those I716065 and I716066 libraries.
- Grew up 9106, 9104, 9203, and flu/inv so I can make more of the mini preps.
- Ordered the hemoglobin and hemoglobin HRP conjugated primary antibody:
http://www.sigmaaldrich.com/catalog/search/ProductDetail/SIGMA/H7379
http://www.bethyl.com/abdetails.asp?antibodyid=967
- Grew up 14 clones from the I716061 plate for the SDS-PAGE gel we're hopefully going to do tomorrow.
- Also sequenced 2 more clones of Chris' 1103-1154 part.
AustinDay 14:30, 18 June 2007 (EDT)
- I digested, ligated, and transformed the 2X mutants (I716065 and I716066)
- I grew up some more of the 1154 clones chris made the other day. The sequences weren't that nice, and one of them wouldn't come out in a digest gel.
Goals for tomorrow:
- Grow up 2X mutant transformants in the morning.
- Use the 1154 and 1150 parts and put em together.
AustinDay 21:09, 14 June 2007 (EDT)
- I minied 2 of the Bca1154 and bca1150 parts that Chris made. I also minied 4 clones of the I716061 library (#2 looked the darkest, and #4 mini was diluted in 100 instead of 50ul.) I also minied 3 clones of the mutant hemoglobins. These mutations should finish up the presbyterian and providence mutation sets. I also sequenced all of them.
AustinDay 18:35, 13 June 2007 (EDT)
- So I finished up the P1 and K1E assay and tried the serum resistance assay today for the talk on saturday.
- I also grew up a few of the I716061 colonies. (They were the ones that should have the cassette with the P-trc-rbs-beta-rbs-alpha) They weren't red, but colony PCR confirmed that they have the correct size. I'm going to let the plate grow up some more and I'm going to grow up a few of them to sequence just to verify the sequence. I'm also going to look into figuring out that spec method of determining the concentration of Hb present.
- I also picked 2 of chris' plates which have the pcon-rbs-alpha and pcon-rbs-beta.
- Here's the pictures of the colony PCR thing I did for the I716061 colonies. The expected band size is 1182 bp.
AustinDay 20:06, 12 June 2007 (EDT)
- Yesterday I transformed the pTRC-rbs-beta-rbs-alpha, but I stupidly choose the wrong restriction sites. Let's just leave that behind us. Anyway, I retransformed those those today, so hopefully we'll see bloody colonies tomorrow.
- I also did the quickchange for the second set of mutations. I plated those today.
- Note to self: Check Chris' plate for hemoglobin colonies (little red ones would be ideal) And put that large taped stack of plates into the mini fridge.
- I colonies appear, pick some and grow em up. Will want to put in tandem like the format I'm working on if they work well.
AustinDay 16:16, 9 June 2007 (EDT)
- I grew up 3 SODs and 3 Met-AP genes for Vai. I also put a bunch of plates into the fridge for some other peoples.
- I minied 3 clones for the 2 hemoglobin subunit and sequenced them. I also minied the antibiotic-rbs library-hemoglobin subunit (unmutated) and digested them so that I can make that antibiotic-rbs-beta-rbs-alpha cassette.
- I also minied the 9106 plasmid of Chris' because the other one I was using for the assays for the symposium data was running low. Those strains are having a hard time transforming. I still can't get any colonies to take up the flu/inv plasmid. I think it might be running low also. Not good.
- I forgot what kind of media I grew up the p-tric transformants into yesterday. But I minied those also and made a digest before I realized that it might have been plain LB. I took a -80 stock of it and grew it up in AMP just in case.
- I also transformed more of the 9104, 9106, flu/inv, and 9023 plasmids into DH10B. Just in case I get a low supply, considering that all of the flu/inv transformations failed last time.
AustinDay 13:43, 8 June 2007 (EDT)
- The transformations of the I716056 and I716057 worked!!! I scraped em and started some cultures.
- The first quickchange reaction seemed to work. I got some colonies. I still went forward with the second quickchange reaction in case somehow these aren't the right results. I grew up 3 of each.
AustinDay 20:02, 7 June 2007 (EDT)
- Redid the digestion and ligations and transformations from yesterday. Which was remaking the I716055 and I716056 (alpha and beta subunits, unmutated, in front of the RBs library and different antibiotics)
- I also realized that I added too much oligo to my quickchange reaction yesterday, so the transformation I did from that may not have worked. I'm redoing the PCR today and running it overnight. Hopefully either the one from yesterday would work, or the one I started again today will work.
AustinDay 15:12, 6 June 2007 (EDT)
- The transformations all ... failed. Not sure why, but I did a lot of them yesterday and they all didn't really work. I'm guessing it's something that was consistently wrong, so I'm going to double check all the temperatures and reagents and try them again. I did the transformation this morning, so hopefully I can get some colonies and grow up the library tonight so that I don't really waste a day because of this.
AustinDay 15:17, 5 June 2007 (EDT)
- Designed the oligos for the hemoglobin mutants. Here are my notes on the mutants:
Primary Mutants:
Wild Type Hemoglobin
PPDA: P50 = 44, yield = 17.5%
Presbyterian mutation (Increase P50): Beta: Asn-108 -> Lys
Providence mutation (Increase yield): Beta: Lys-82 -> Asp
Di-alpha fusion (Increase yeidl): Glycine? linker.
P2996: P50 = 34, yield “average”
Presbyterian mutation (Increase P50): Beta: Asn-108 -> Lys
αL29F (Decrease P50, decrease auto-oxidation)
αV96W (Increase P50, increase auto-oxidation)
Oligo design for the following mutations:
ASN: AAC -> AAA
B-Asn108Lys
Lys AAG 14043.00 10.26 0.23
Lys AAA 46044.00 33.66 0.77
GAATTTTCGTCTGCTGGGTAAAGTGCTGGTGTGTGTCCTTGC
LYS: AAA -> GAT
B-Lys82Asp
Asp GAT 44103.00 32.24 0.63
Asp GAC 26201.00 19.15 0.37
CTGGCGCATCTTGATAATCTTGATGGTACATTCGCGACTCTGAG
Di-alpha fusion: Oligos coming soon.
LYS: TTA -> TTT
A-L29F
Phe TTT 30361.00 22.19 0.57
Phe TTC 22649.00 16.56 0.43
GAGTATGGTGCTGAAGCTTTTGAACGCATGTTTTTAAGCTTTC
VAL: GTT -> TGG
A-V96W
Trp TGG 20835.00 15.23 1.00
CGCATAAACTCCGTGTGGACCCGTGGAACTTTAAACTGCTGTCCCACTG
Secondary Mutants (Or mutations that would lead to them)
Alpha; VAL E11 -> Threonine: Increases P50 by 4x (to 25 torr). Due to stabilization of water molecule in distal heme pocket of decoy T-state. Increases auto-oxidation rate.
Beta; VAL E11 -> Threonine: Increases P50 by 2x (to 12 torr).
Alpha; V26T: 4x increase in P50. Lost cooperativity. (n = 1.1)
Beta; B67T: (n = 2.2) Slightly increased P50.
Combination of surface modifications to increase P50 by 3x:
o Val1 -> Met:
o His2 -> deleted
o Thr4 -> Ile
o Pro5 -> Ala
o Ala 76 -> Lys
βN108D, (In place of presbyterian) increases the P50 more than the presbyterian.
- In other news: All but I716053 #1 came out white. That colony was red, so I didn't mini it. (parent vector)
- I'm going to go on with the construction now with the RBs libraries from yesterday.
- I used the #1 clone from each of the parts for the next digest/ligation/transformation round.
- I transformed the colonies with the RBS library. I forgot to select for the secondary antibiotic, but Chris said I should just scrape the plate tomorrow and grow it all up in the antibiotic marker. Hopefully that'll do the trick.
AustinDay 13:25, 4 June 2007 (EDT)
- Got a nice number of colonies on the synthetic gene biobrick transformations from yesterday. I grew up 2 of each.
- I minied the RBS library parts (I716051 and I716052).
AustinDay 16:50, 3 June 2007 (EDT)
- Made an EcoRI / XhoI digest of the synthetic genes and a digest of EcoRI and XhoI of the 9145 plasmid.
- Here's the gel. Very pretty.
- Took the small fragment from the synthetic genes and the larger fragment from the 9145 plasmid and ligated them together.
- I think I'm going to name these new parts: I716053 (9186 + 9145), I716054 (9187 + 9145), I716055 (9188 + 9145). These names came to me in a dream. It was said they shall bring luck and prosperity to all who use them.
- I grew up 2 colonies of the homogenized RBS pools. (The Kan and the Cmr pools)
Austinday 15:43, 2 June 2007 (PDT)
- Okay, so since the 1121 part was bad, chris found a PCR product of it. I'm going to give that a try and see if it works.
- I did the ligation and transformation for those. I named the parts Bad0001 (kan) and Bad0002 (Cmr). Plated and cooking for tomorrow.
- Made digest of BglII and XhoI of the 3X concentrated 9145 plasmid miniprep. The total digest was equal to that of 14 regular digests. The gel is below:
- Digested the DNA 2.0 plasmids with EcoR1 and XhoI. But...the gel looks funny. I may not have allowed enough time for the lyophilized DNA to redissolve... That's my best guess at least. (Update: Nope, I think I just overloaded the lanes)
- I grew up some colonies of the DNA 2.0 parts so I'll give it another try tomorrow. (Chris told me that there is probably too much DNA and that if I ran it longer, it might have cleared up. You can kind of see that there might be a second band...sort of.)
Austinday 22:59, 30 May 2007 (PDT)
- Okay, I repeated the digestion from yesterday, but got the same results. Here's the gel.
- This time, I let it digest for well over an hour and mixed it half way through. I also ran the gel at a lower voltage than usual.
- The bands for the 1122 lane was noticeably cleaner, but the 1121 lane was uncut.
- I cut out the smaller band from the 1122 lane(because it's probably a higher yield than the previous gel), as well as the 1128 band. I figured that because the previous gel didn't cut completely, and because the 1128 pool smaller bands would have already ran off the gel, this second gel would have a more pure band than the first. I also cut out the region of the 1121 lane where the smaller band should be, although it looks as if nothing is there... I guess we'll have to remake that 1121 part.
Austinday 16:53, 29 May 2007 (PDT)
- Sequencing of the pBca1101-Bca1128 RBS library resulted in 2 duplicate RBS's. #24 was identical to #3, and #5 was identical to #2 so I threw out #3 and #5.
- I pooled together the bca1128 rbs's and the 1106 A and B rbs's as well as one other 1128 A rbs Chris had.
- I digested those with EcoR1 and BglII to create the vectors to insert the antibiotic markers. After digestion, the pool looked good so I cut it out right away. The 1121 and 1122 (CmR and Kan) digestions didn't look correct.
- The lanes are: Marker (Barely came out), 1121, 1122, and rbs pool (cut out)
- 1121 has the right number of bands, but I can't exactly tell if the size of the smaller band is correct.
- 1122 has the wrong number of bands. I'm not that experienced with picking out when a band is undigested plasmid, but even if that were the case for the third band, the smallest band looks much too small.
- Anyway, I cut out the smallest bands from 1121 and 1122 and stored them for tomorrow. Maybe Chris will have something to add to my gelatinous adventures.
198.128.27.101 13:55, 25 May 2007 (PDT)
- Preliminary ranking of RBS's picked from the library based on culture colors: (Bca1101 - Bca1128's in TG1)
Stronger --> Weaker
24, 1, 10, 18, 2, 3, 5, 9, 19, 4, 12, 11, 20, 21, 16, 13, 17, 7, 6, 15, 22, 14, 23
The first 7 are significantly more red than the rest.
# 11 was lost due to a tragic mini prepping accident. May it rest in peace.
AustinDay 16:38, 3 June 2007 (EDT)
- Time to start some awesomeness.
- Biobrick numbers: 051-100 Austin Day
- The next few entries are cut and pasted from my Arkin Wiki because I started making entries on that before this wiki was set up.