Edinburgh/DivisionPopper/Applications
From 2007.igem.org
(→Counting using more recombination sections) |
|||
| Line 22: | Line 22: | ||
===Counting using more recombination sections=== | ===Counting using more recombination sections=== | ||
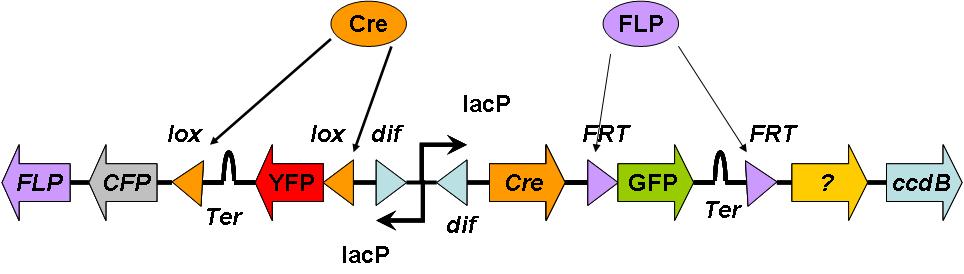
| - | Rather than using the DivisionPoPper directly, this uses the flipping dif sites to activate the different recombination sites and cut out the | + | Rather than using the DivisionPoPper directly, this uses the flipping dif sites to activate the different recombination sites and cut out the sections of DNA |
[[Image:Cell division.jpg|800px]] | [[Image:Cell division.jpg|800px]] | ||
Revision as of 11:04, 6 August 2007
Edinburgh > DivisionPopper
Introduction | Applications | Design | References
https://static.igem.org/mediawiki/2007/f/f5/800px-Edinburgh_City_15_mod.JPG
Applications and further research
Contents |
This page details some of the potential uses of the Division PoPper and other ways of using the recombination systems
Division Frequency Analysis
The output of the Division PoPper could be linked to the production of a slowly degrading protein. The more frequent the divisions, the grater the concentration of the protein.
Division Counting
Coupling to a PoPS counting device
Couple the output of the Division PoPper to another counting device (such as the ETH Zurich counter or other variants) to count the number of cell divisions. This would be difficult to test due to the nature of colonies and the cells dividing out of phase
Counting using more recombination sections
Rather than using the DivisionPoPper directly, this uses the flipping dif sites to activate the different recombination sites and cut out the sections of DNA