Experiments
From 2007.igem.org
(Difference between revisions)
(→Open loops) |
(→Experiment) |
||
| Line 31: | Line 31: | ||
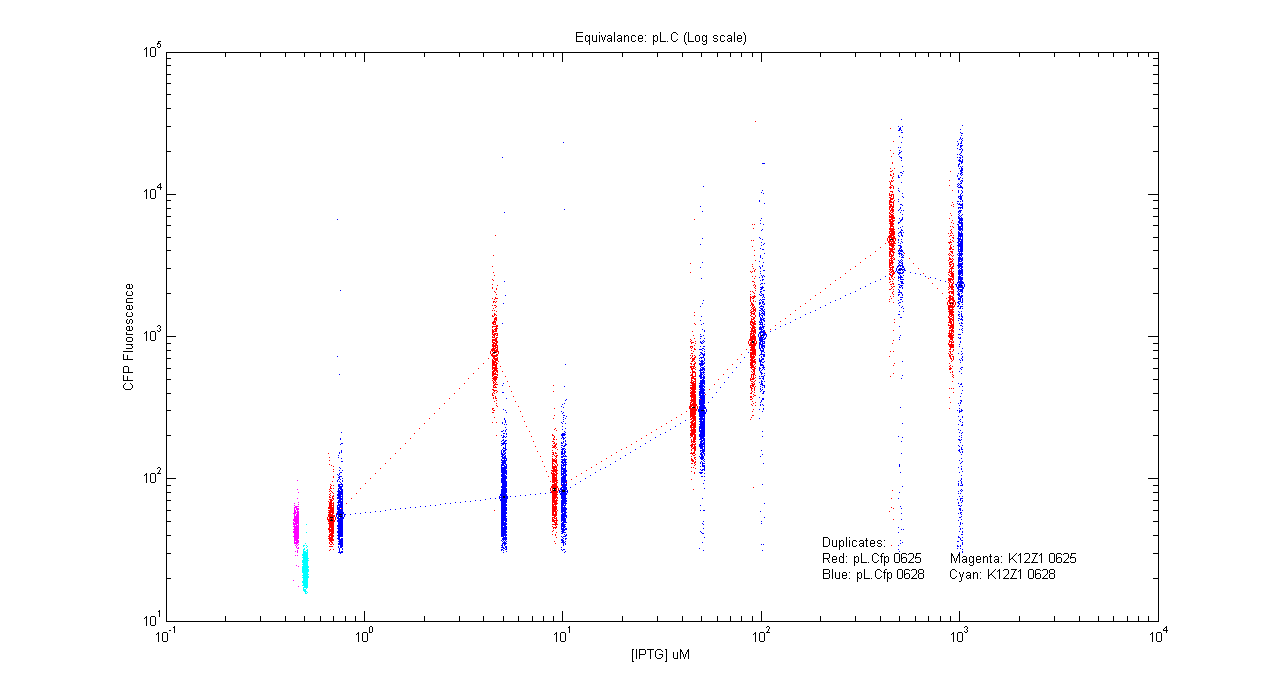
*pL.Cfp | *pL.Cfp | ||
| - | [[Image:pLC_dup.png]] | + | [[Image:pLC_dup.png|400px]] |
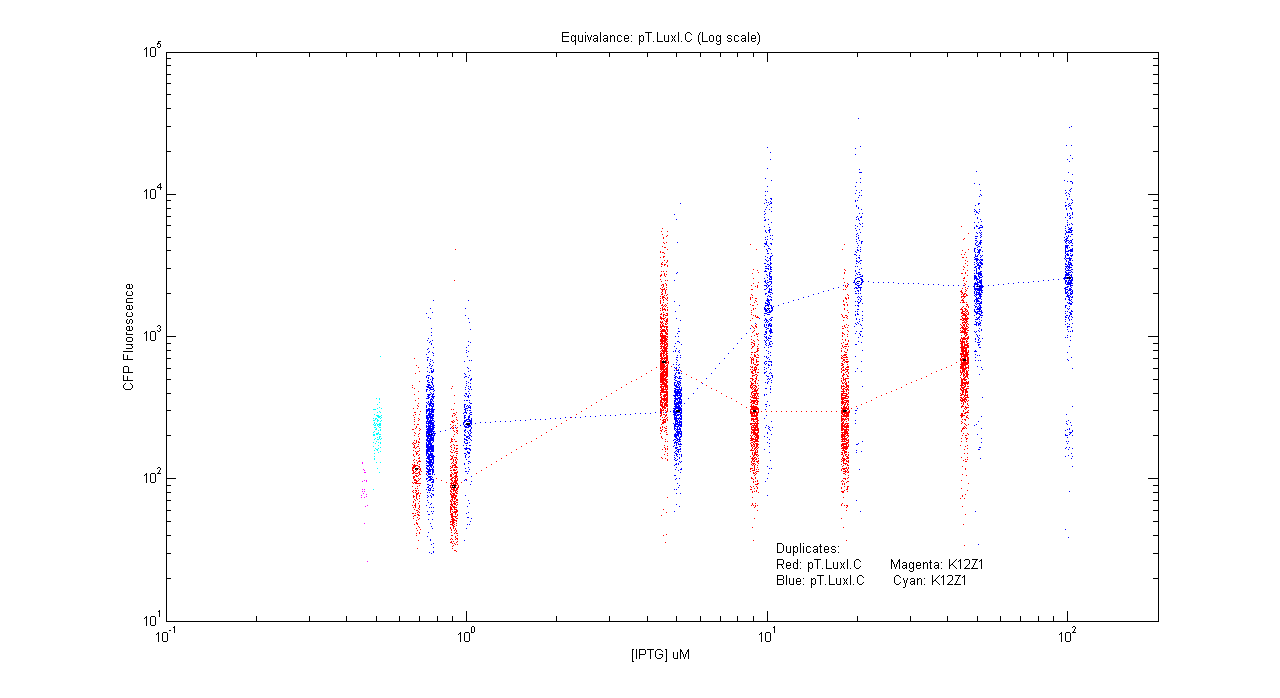
*pT.luxI.Cfp | *pT.luxI.Cfp | ||
| - | [[Image:pTIC_dup.png]] | + | [[Image:pTIC_dup.png|400px]] |
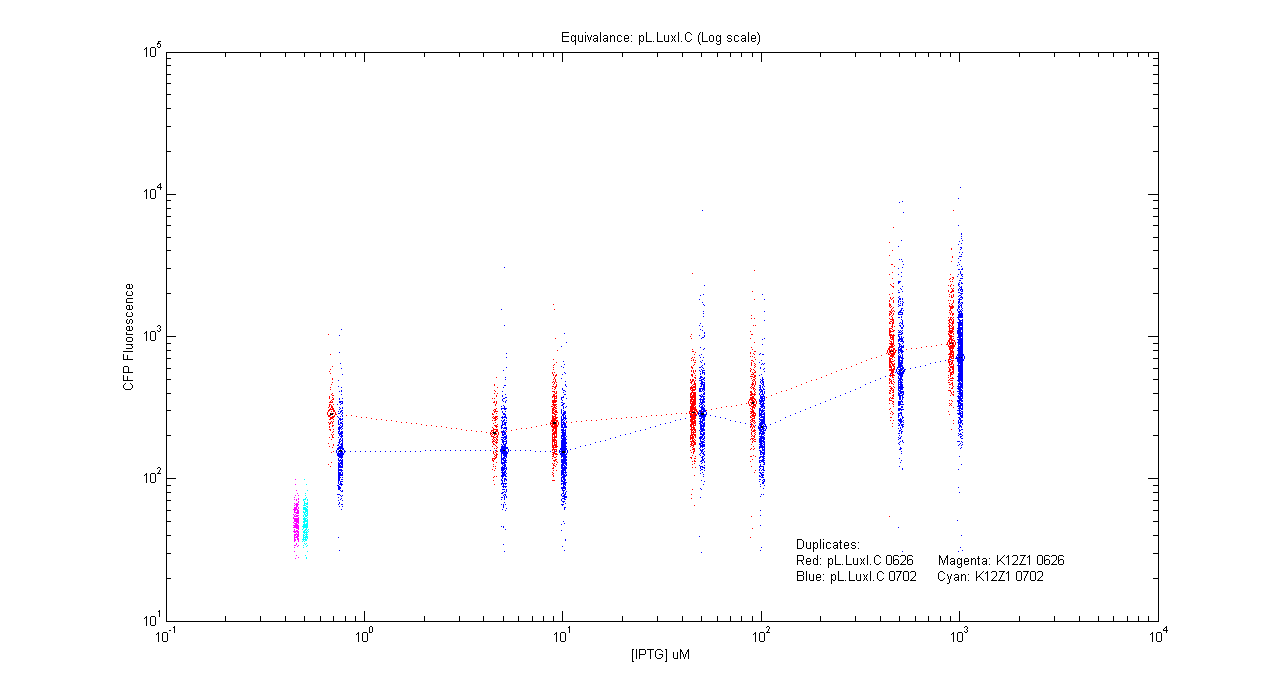
*pL.luxI.Cfp | *pL.luxI.Cfp | ||
| - | [[Image:pLIC_dup.png]] | + | [[Image:pLIC_dup.png|400px]] |
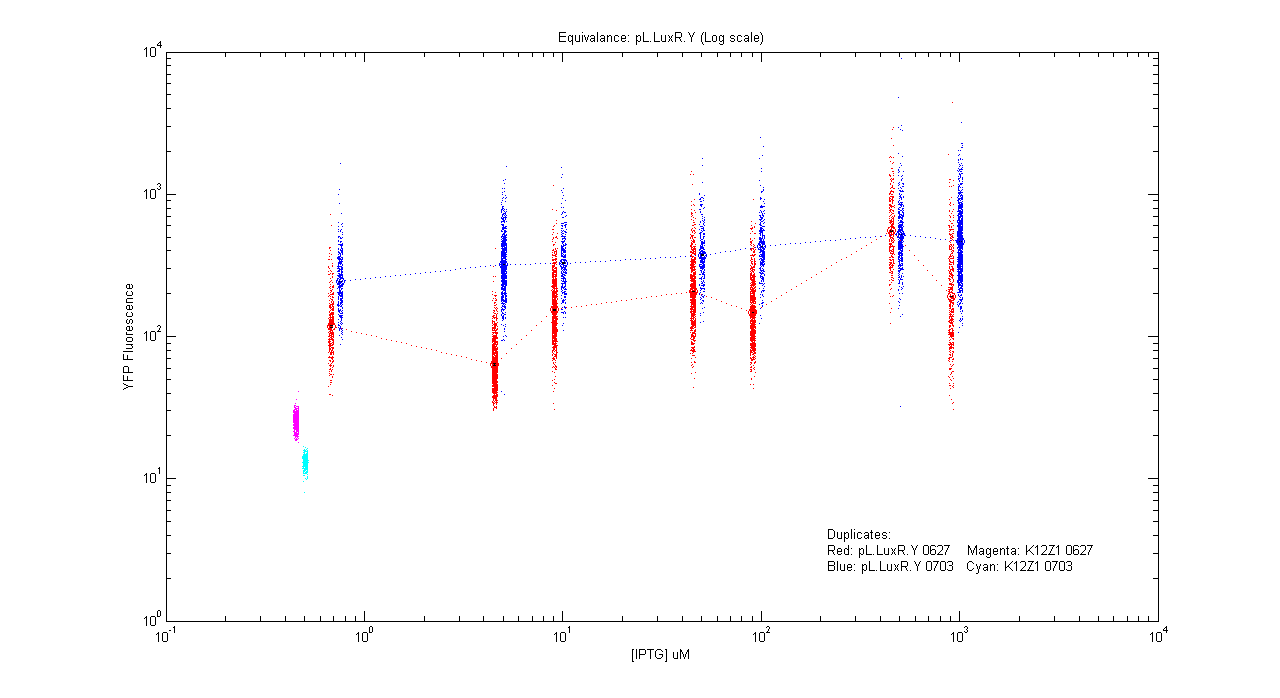
*pL.luxR.Yfp | *pL.luxR.Yfp | ||
| - | [[Image:pLRY_dup.png]] | + | [[Image:pLRY_dup.png|400px]] |
=== Open loops === | === Open loops === | ||
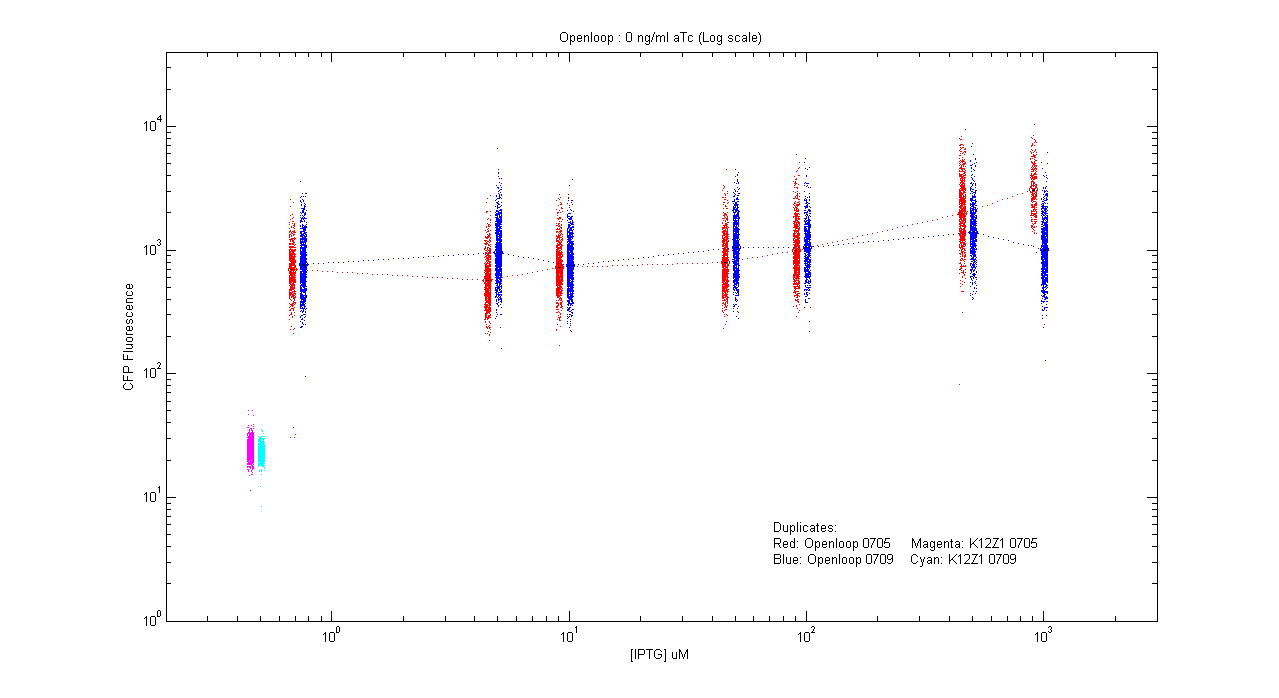
*pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (0 ng/ml aTc) | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (0 ng/ml aTc) | ||
| - | [[Image:Openloop_0.png]] | + | [[Image:Openloop_0.png|400px]] |
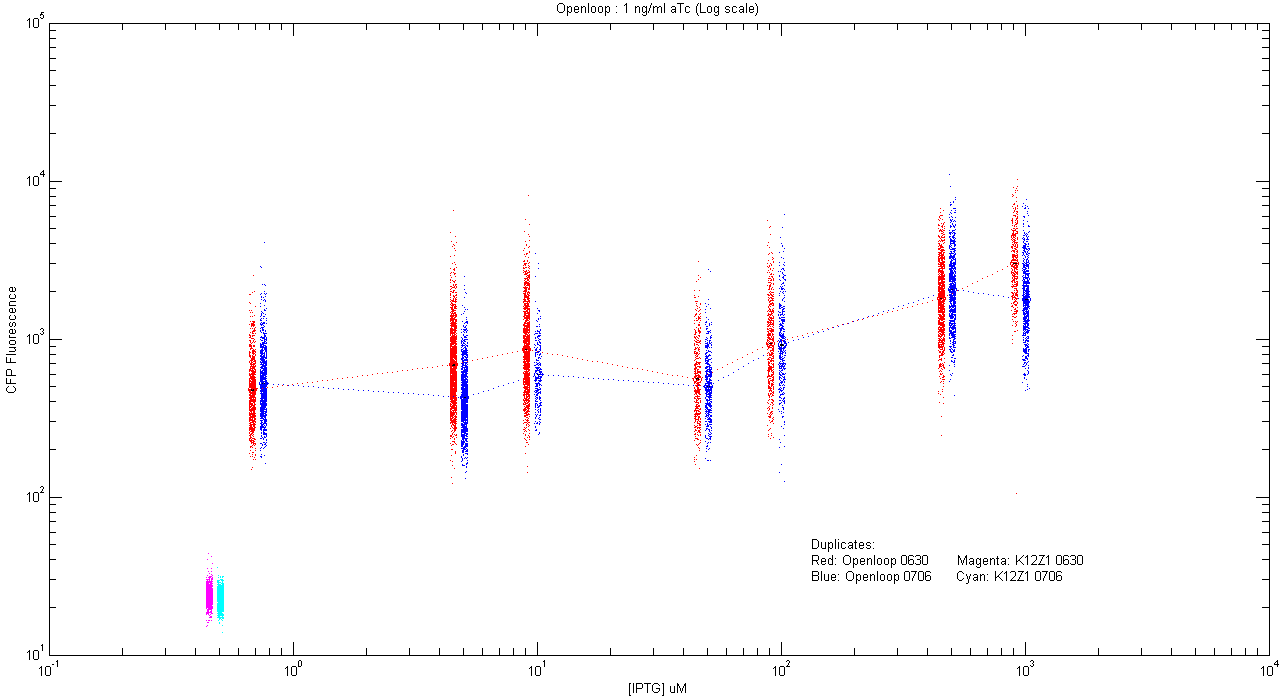
*pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (1 ng/ml aTc) | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (1 ng/ml aTc) | ||
| - | [[Image:Openloop_1.png]] | + | [[Image:Openloop_1.png|400px]] |
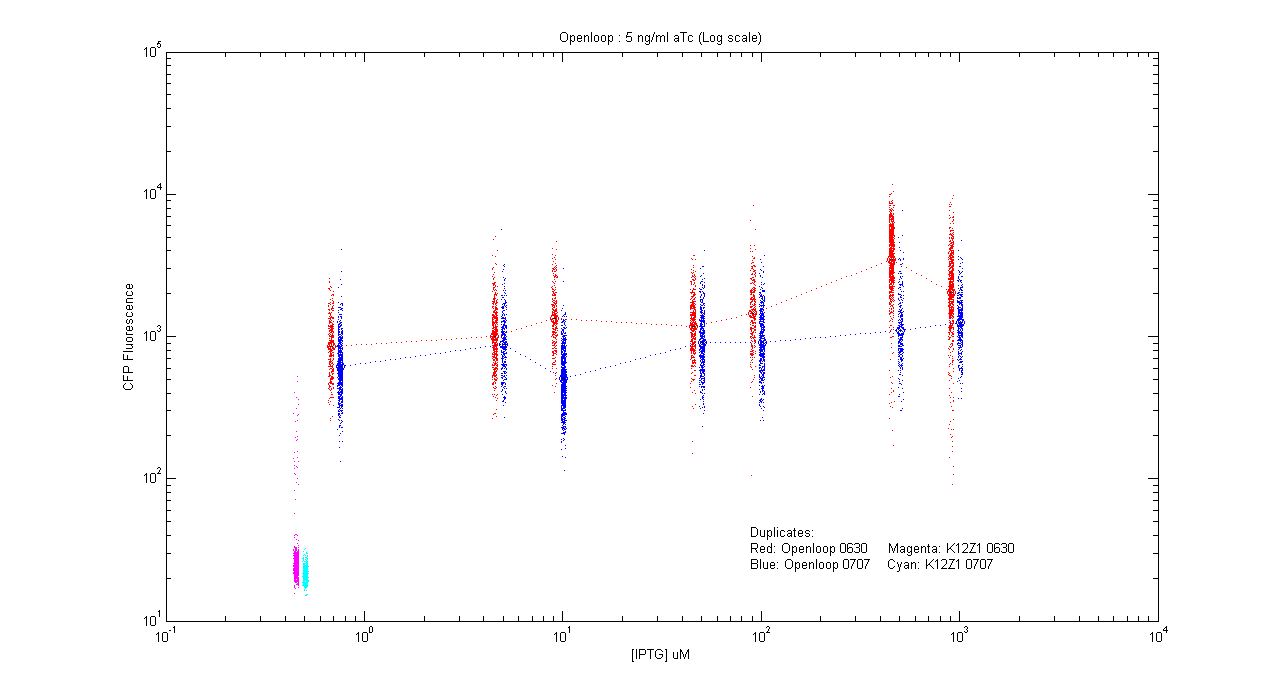
*pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (5 ng/ml aTc) | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (5 ng/ml aTc) | ||
| - | [[Image:Openloop_5.png]] | + | [[Image:Openloop_5.png|400px]] |
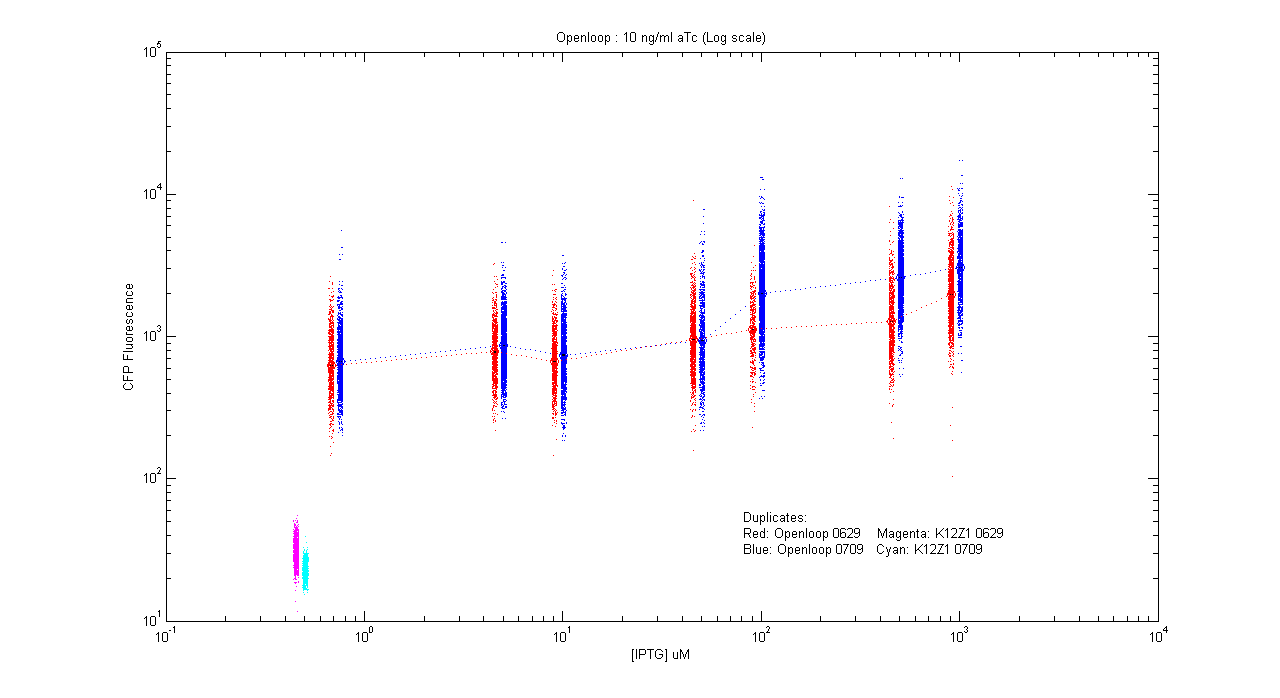
*pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (10 ng/ml aTc) | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (10 ng/ml aTc) | ||
| - | [[Image:Openloop_10.png]] | + | [[Image:Openloop_10.png|400px]] |
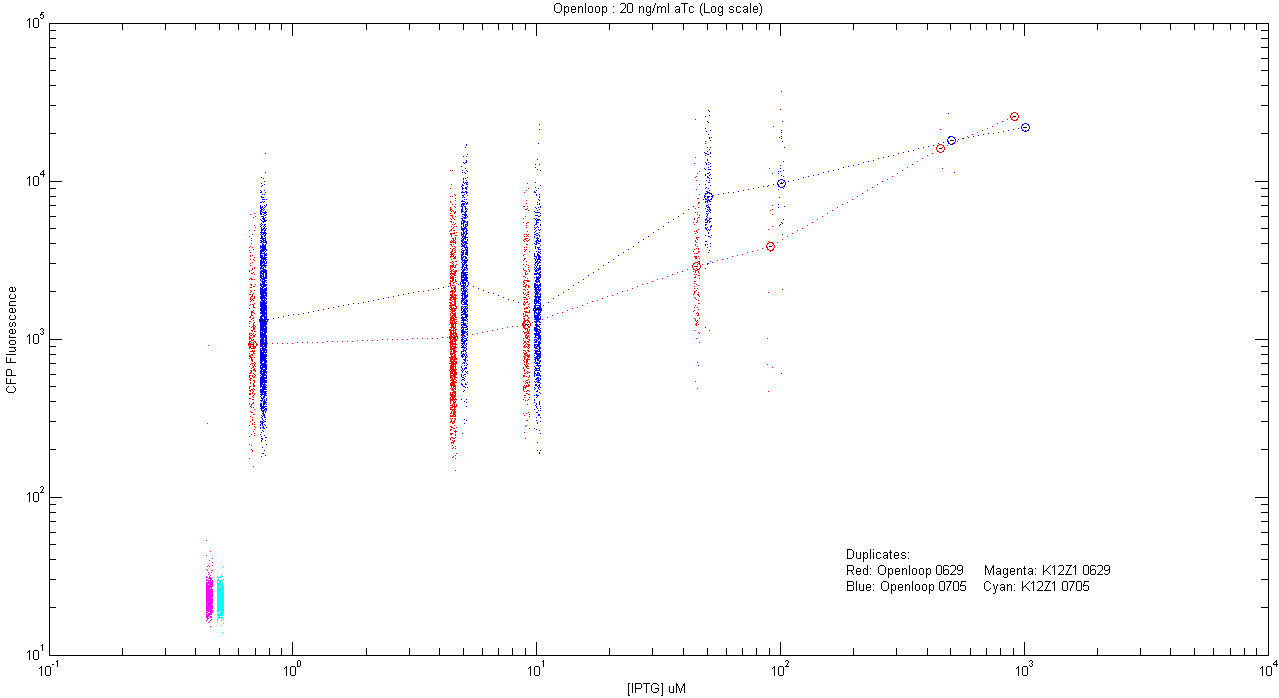
*pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (20 ng/ml aTc) | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (20 ng/ml aTc) | ||
| - | [[Image:Openloop_20.png]] | + | [[Image:Openloop_20.png|400px]] |
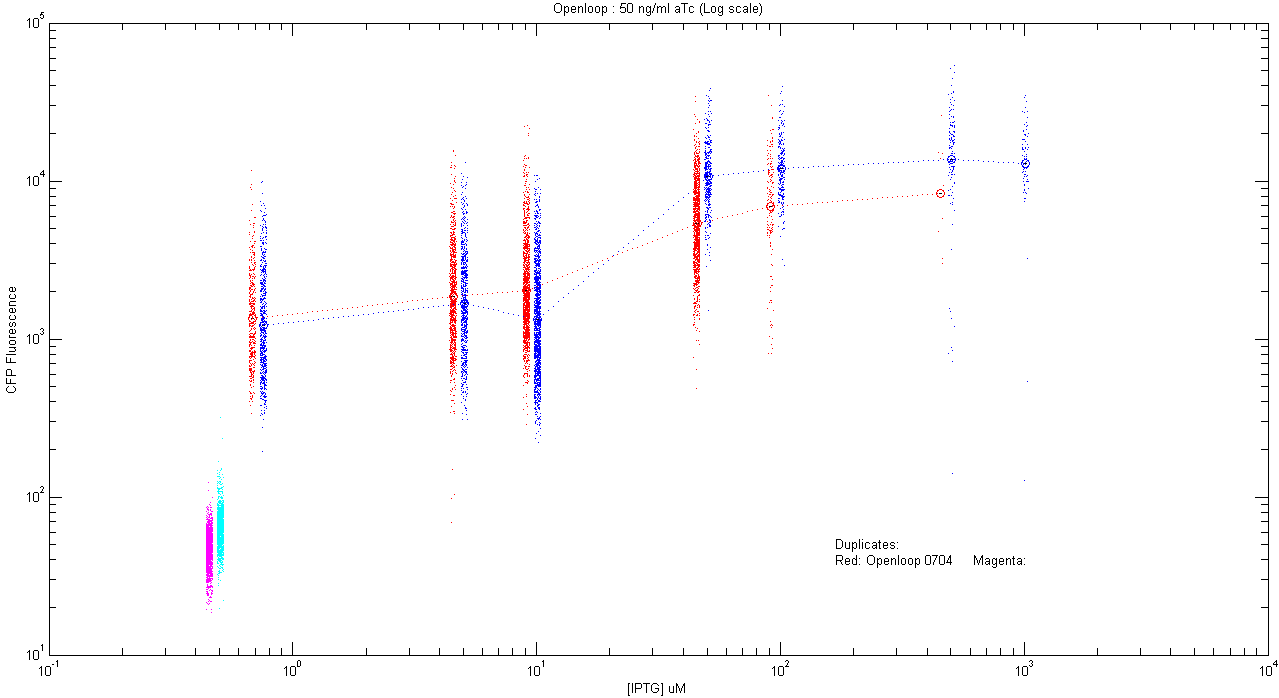
*pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (50 ng/ml aTc) | *pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (50 ng/ml aTc) | ||
| - | [[Image:Openloop_50.png]] | + | [[Image:Openloop_50.png|400px]] |
=== Closed loops === | === Closed loops === | ||
Revision as of 15:47, 24 August 2007
| The official wiki of the NCBS iGEM 2007 Team | |
| The NCBS iGEM 2007 Experiments | |
|
The following is the record of all the experiments done by us, each followed by graphs obtained by analysis of the corresponding microscopy and flow cytometry data. Note that in flow cytometry a signal obtained from a filter does not exactly correspond to CFP or YFP amount inside a cell; when cells express both the proteins. This is because of spectral overlap of their excitation and emission spectra. We came up with a mathematical method to separate CFP and YFP from autofluorescence and noise. Click here for details about this mathematical tool for correction. |
| [http://www.ncbs.res.in/ National Centre for Biological Sciences, Bangalore] |
|---|
| Bangalore | The Team | The Mission | Experiments | e-Notebook |
|---|
Contents |
Experiment
Equivalences
- pL.Cfp
- pT.luxI.Cfp
- pL.luxI.Cfp
- pL.luxR.Yfp
Open loops
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (0 ng/ml aTc)
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (1 ng/ml aTc)
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (5 ng/ml aTc)
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (10 ng/ml aTc)
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (20 ng/ml aTc)
- pT.luxI.Cfp::pL.luxR.Yfp.pR.Cfp (50 ng/ml aTc)