Virginia/Projects/2
From 2007.igem.org
| Line 15: | Line 15: | ||
==Harvesting Biomass and Light to Power Butanol Biosynthesis== | ==Harvesting Biomass and Light to Power Butanol Biosynthesis== | ||
| + | |||
| + | At the 2007 iGEM Jamboree, we will present our synthetic biology approach to the design and implementation of a novel metabolic pathway in ''E. coli'' that utilizes cellulose and sunlight as sole energy sources for the biosynthesis of butanol, an alternative liquid transportation fuel. Click [http://www.openwetware.org/wiki/IGEM:VGEM/2007/Projects here] to find our more about our other projects that will not be presented this year. | ||
===Methods and Materials=== | ===Methods and Materials=== | ||
Revision as of 19:23, 16 September 2007
| HOME | PROJECT INTRO | APPROACH | PROCEDURES | RESULTS | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2007&group=Virginia BIOBRICKS] | [http://openwetware.org/wiki/IGEM:VGEM/2007/Notebook eNOTEBOOK] | [http://www.seas.virginia.edu/VGEM/ WEBSITE] |
Harvesting Biomass and Light to Power Butanol Biosynthesis
At the 2007 iGEM Jamboree, we will present our synthetic biology approach to the design and implementation of a novel metabolic pathway in E. coli that utilizes cellulose and sunlight as sole energy sources for the biosynthesis of butanol, an alternative liquid transportation fuel. Click [http://www.openwetware.org/wiki/IGEM:VGEM/2007/Projects here] to find our more about our other projects that will not be presented this year.
Methods and Materials
We will analyze butanol production via gas chromatography and mass spectroscopy.
Our experimental scaffold (still in development) is as follows:
What is the butanol tolerance of WT cells and of transformed cells?
1) Butanol Tolerance
Hypothesis: Transformed cells will be more resistant to extracellular butanol than WT
* Add varying amounts of butanol to broths of transformed and non-transformed cells * Determine amounts of butanol cells can withstand before dying * Count cells with hemocytometer and blue stain
Note: Depending on butanol tolerance we will consider butanol tolerance transformed cells as wildtype (WT). If it doesn't give any significant tolerance we will just use the original strain.
Can cells survive with only cellulose carbon source?
2) Effect of Cellulase
Hypothesis: Transformed cells should be able to grow on low glucose/high cellulose media. No growth for WT.
* Transform cells with cellulase * Grow transformed cells and control cells on low glucose media supplemented with cellulose * Observe growth amounts
Does thiolase increase the production of acetoacetyl-CoA from acetyl-CoA relative to WT?
3) Effect of Thiolase
Hypothesis: Thiolase-transformed cells will have increased acetoacetyl-CoA production than WT, and most if cellulase-transformed
* Transform cells with thiolase * Transform cellulase-cells (ex.2) with thiolase * Plate transformed cells (thiolase and thiolase + cellualse, and control cells) * Test for acetoacetyl-CoA production
Note: If thiolase confers much greater acetoacetyl-CoA production, we will include it by default in the rest of the experiments requiring the central pathway.
How much butanol is produced from acetyl coa to butanol, with and without alcohol dehydrogenase?
4a) Acetyl CoA to Butanol (-cellulase, +thiolase, +central pathway, +aad, -alcohol dehydrogenase, -AOTC/AOTD)
Hypothesis: Transformed cells will produce butanol. WT will not
* Transform cells with thiolase, central pathway, aad/aad2 * Grow transformed and control cells * Test for Butanol
4b) Acetyl CoA to Butanol (-cellulase, +thiolase, +central pathway, +aad, +alcohol dehydrogenase, -AOTC/AOTD)
Hypothesis: Transformed cells will produce butanol, more than in ex. 4a. WT will not
* Transform cells with thiolase, central pathway, aad/aad2, alcohol dehydrogenase * Test for butanol production
How much butanol is produced from cellulase to acetyl coa to butanol, with and without alcohol dehydrogenase?
5a)Cellulase to Acetyl CoA to Butanol (+cellulase, +thiolase, +central pathway, +aad, -alcohol dehydrogenase, -AOTC/AOTD)
Hypothesis: Transformed cells will produce butanol. WT will not.
* Transform cells with cellulase, thiolase, central pathway, and aad/aad2 * Grow transformed and control cells * Test for Butanol
5b) Cellulase to Acetyl CoA to Butanol (+cellulase, +thiolase, +central pathway, +aad, +alcohol dehydrogenase, -AOTC/AOTD)
Hypothesis: Transformed cells will produce butanol, and at higher amounts than in ex. 5a. WT will not.
* Transform cells with cellulase, thiolase, central pathway, aad/aad2, alcohol dehydrogenase * Grow transformed and control cells * Test for Butanol
How much butanol is produced via only the bottom pathway (AOTC/AOTD genes) with and without the alcohol dehydrogenase?
6a)Butyrate (butyric acid) to Butanol (-cellulase, -thiolase, -central pathway, +aad, -alcohol dehydrogenase, +AOTC/AOTD) IN FLOW
Hypothesis: Pathway will produce butanol. There will be no butanol in WT
* Transform cells with AOTC/AOTD and aad/aad2 * Grow transformed and control cells * Measure butanol production with varying concentrations of initial butyric acid
6b) Butyrate (butyric acid) to Butanol (-cellulase, -thiolase, -central pathway, +aad, +alcohol dehydrogenase, +AOTC/AOTD)
Hypothesis: Pathway will produce butanol, more than in ex.6a. There will be no butanol in WT
* Transform cells with aad, AOTC/AOTD, and alc. dehydro. * Grow transformed and control cells * Measure butanol production with varying concentrations of initial butyric acid
How much butanol from acetyl coa to butanol, including AOTC/AOTD, and with and without alcohol dehydrogenase?
7a) Acetyl CoA to Butanol with Butyrate pathway (-cellulase, +thiolase, +central pathway, +aad, -alcohol dehydrogenase, +AOTC/AOTD)
Hypothesis: This is the complete pathway, but without alcohol dehyd. High butanol production expected.
* Transform cells with thiolase, central pathway, aad, AOTC/AOTD * Test for butanol production
7b) Acetyl CoA to Butanol with Butyrate pathway (-cellulase, +thiolase, +central pathway, +aad, +alcohol dehydrogenase, +AOTC/AOTD)
Hypothesis: This is the complete pathway, without cellulase. High butanol production expected.
* Transform cells with thiolase, central pathway, aad, AOTC/AOTD, and alc. dehydro. * Test for butanol production
How much butanol from cellulase to acetyl coa to butanol, including ctfa/b, and with and without alcohol dehydrogenase?
8a) Cellulase to Butanol with Butyrate pathway (+cellulase, +thiolase, +central pathway, +aad, -alcohol dehydrogenase, +AOTC/AOTD)
Hypothesis: This is the complete pathway, without alcohol dehydr. There should be maximum production of butanol.
* Transform cells with cellulase, thiolase, central pathway, aad, AOTC/AOTD, and alc. dehydro. * Test for butanol production
8b) Cellulase to Butanol with Butyrate pathway (+cellulase, +thiolase, +central pathway, +aad, +alcohol dehydrogenase, +AOTC/AOTD)
Hypothesis: This is the complete pathway. There should be maximum production of butanol.
* Transform cells with cellulase, thiolase, central pathway, aad, AOTC/AOTD, and alc. dehydro. * Test for butanol production
Will cells transformed with the proteorhodopsin pathway survive with only light, will it produce butanol, and if so how much will it increase butanol production when combined with other pathways?
9a) Effect of Proteorhodopsin
* Transform cells with proteorhodopsin genes * grow on minimal nutrient media * observe growth vs. control strains
9b) Proteorhodopsin to butanol
* If ex.9a cells grow at all, * transform cells with central pathway genes and proteorhodopsin * test for butanol production
9c) Proteorhodopsin to butanol, full pathway
* add proteorhodopsin to ex.8b cells * test for butanol production
Results and Conclusions
TBD
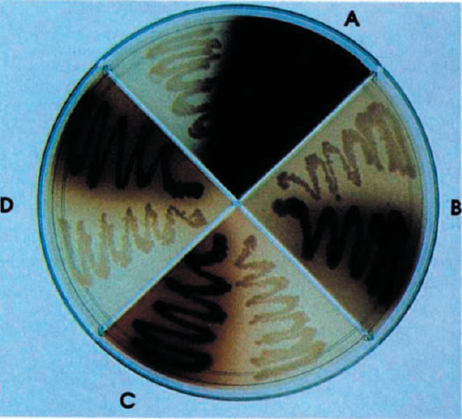
Bacterial Melanogenesis
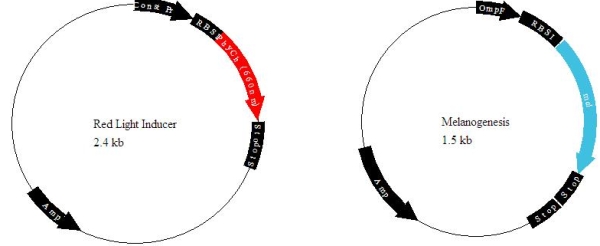
We have designed a light-inducible system in E. coli capable of manufacturing and secreting melanin. This biological machine is built from two standard biological parts, a melanin generator and a red light-inducible plasmid.
Motivation
According to the American Cancer Society, skin cancer is the most common form of all cancers. It accounts for nearly half of all cancers in the United States. Due to its natural defense mechanism, skin must first be exposed to UV radiation before melanogenesis begins. Thus, we thought it would be convienient to be able to produce melanin in response to another harmless wavelength. This technology may, in the future, lead to treatments for albinism and a safe alternative for cosmetic tanning. Melanin may also be used as a natural indicator in the laboratory as well as an acoustic material. According to a study by Kono, et. al., melanin is capable of absorbing ultrasound.
Additionally, we would like to be able to use the melanin secretion system as an indicator in our ethylene detection project. Theoretically, more and more melanin would be secreted as various produce items become riper and produce more ethylene, causing the indicator to become darker on a gradient.
Background
In humans, melanin is found in the skin, hair, iris, adrenal gland, inner ear, and various pigment-producing regions of the brain.
Melanin is the primary determinant of skin color. The process of tanning originates in specialized cells called melanocytes. When stimulated by ultraviolet radiation, these cells produce melanin, which then migrates to the neighboring keratinocytes. The melanin accumulates above the receiving cells' nuclei in order to protect genetic material from from mutations caused by the sun's ionizing radiation.
Though there are many types of melanin, they are all polymers which contain derivatives of the amino acid tyrosine. Melanin is an aggregate of smaller component molecules; the various types of melanin have differing proportions and bonding patterns of the component molecules. Three types of melanin found in humans include eumelanin, pheomelanin, and neuromelanin. Eumelanin, which produces a brown-black color, is most common.
References:
Wu, Corrina. Unraveling the Mystery of Melanin. Sept 18, 1999. Vol 156, No 12.
[http://www.mbceo.com/science/more_information.php?c=Melanin Information and Facts About Melanin]
Biobrick design
This project has a very simple design: the melanin producing gene mel from Streptomyces is linked to the red-light inducer designed by UT Austin in the iGem 2006 competition.
Methods and Materials
methods of measurement
Results and Conclusions
TBD
Send comments to George McArthur