Paris/October 1
From 2007.igem.org
(Difference between revisions)
(→pDapA DAP-dependant repression through FACS analysis) |
|||
| Line 21: | Line 21: | ||
== pDapA DAP-dependant repression through FACS analysis == | == pDapA DAP-dependant repression through FACS analysis == | ||
| + | |||
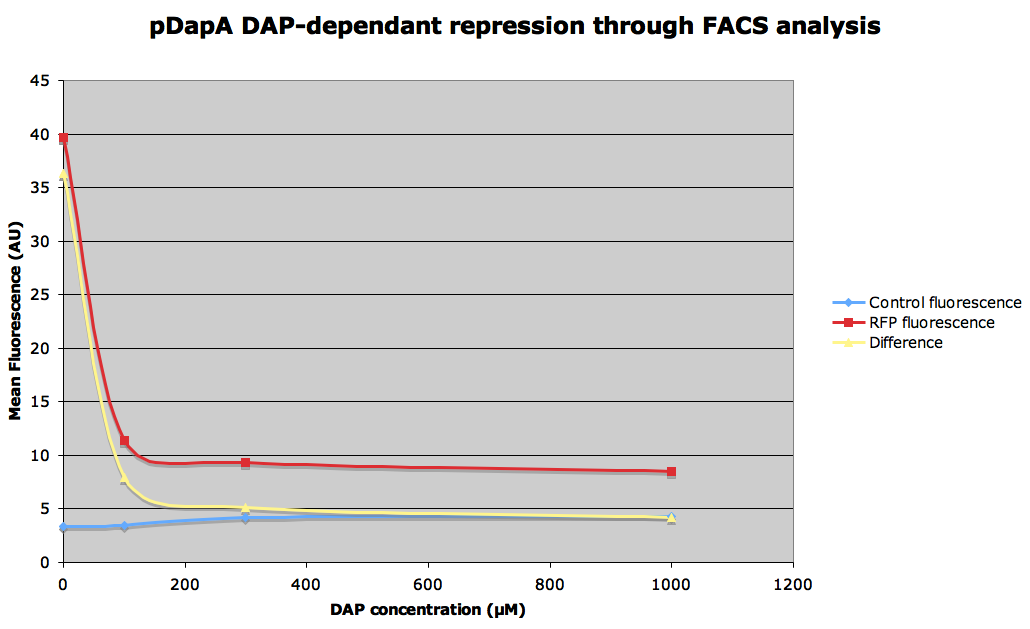
| + | Strain FR781 containing a plasmid with the mRFP expression controlled by the promotor pDapA has been cultured overnight in 5 mL of LB with different concentrations of DAP.<br> | ||
| + | <br> | ||
| + | - 1mM<br> | ||
| + | - 300 µM<br> | ||
| + | - 100 µM<br> | ||
| + | - 50 µM<br> | ||
| + | <br> | ||
| + | - 0 µM environment has been simulated by dilluting and letting overnight 300µM DAP LB culture at 1/100 in minimal medium M9 without DAP during 1h before analysis at 4°C.<br> | ||
| + | <br> | ||
| + | All analyses are made on 1/100 dillution of overnight LB culture in minimal medium M9 at 4°C.<br> | ||
[[Image:pDapA FACS.jpg|thumb|800px|]] | [[Image:pDapA FACS.jpg|thumb|800px|]] | ||
Revision as of 14:54, 3 October 2007
PCR on Wanner Bfr constructions (L58.5 &L58.7) with the Bfr-Ftsk primers (O43 & O44)
- We used the Phusion high-fidelity DNA Polymerase (Finnzymes) :
- 26,5µL H2O
- 10µL 5X Buffer
- 7µL DNA template
- 2*2,5µL Primers
- 1µL dNTP
- 0,5µL Polymerase
- 2-step PCR reaction : the Tm was higher than 72°C, then Finnzymes asks for us to not use an annealing time anymore.
- 98°C initial denaturation : 1'30
30 steps of :
- 98°C denaturation : 30"
- 72°C elongation : 50"
and
- 72°C final elongation : 7'
pDapA DAP-dependant repression through FACS analysis
Strain FR781 containing a plasmid with the mRFP expression controlled by the promotor pDapA has been cultured overnight in 5 mL of LB with different concentrations of DAP.
- 1mM
- 300 µM
- 100 µM
- 50 µM
- 0 µM environment has been simulated by dilluting and letting overnight 300µM DAP LB culture at 1/100 in minimal medium M9 without DAP during 1h before analysis at 4°C.
All analyses are made on 1/100 dillution of overnight LB culture in minimal medium M9 at 4°C.