ETHZ/Biology
From 2007.igem.org
Stefan Luzi (Talk | contribs) |
Stefan Luzi (Talk | contribs) |
||
| Line 83: | Line 83: | ||
|- | |- | ||
! 2 | ! 2 | ||
| - | | [http://partsregistry.org/Part: | + | | [http://partsregistry.org/Part:BBa_I739102 (cI negative / TetR negative) promoter] (BBa_I739002): inner part of system, no part of the system |
|} | |} | ||
| - | + | <br> | |
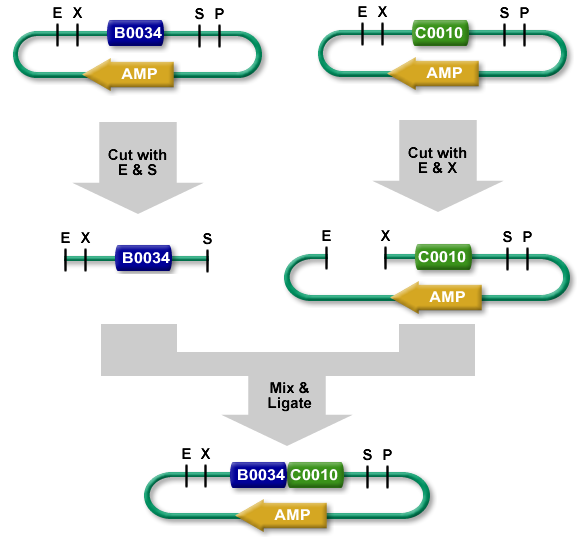
The standard BioBrick assembly will be used to put the parts in the plasmids. Detailed information on how the BioBrick part fabrication works can be found [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication here]. For a shorter explanation of how to assemble two parts together check [http://partsregistry.org/Assembly:Standard_assembly here] (Fig. 4). Note that the composite part is constructed from the end to the beginning, i.e. each new part is inserted ''before'' the existing one. In the following, the plasmid containing the new part to be inserted will be referred to as the ''donor'' and the plasmid accepting the new part will be referred to as the ''acceptor''. Composite pars made of parts '''a''' and '''b''' are denoted '''a.b'''.</p><br> | The standard BioBrick assembly will be used to put the parts in the plasmids. Detailed information on how the BioBrick part fabrication works can be found [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication here]. For a shorter explanation of how to assemble two parts together check [http://partsregistry.org/Assembly:Standard_assembly here] (Fig. 4). Note that the composite part is constructed from the end to the beginning, i.e. each new part is inserted ''before'' the existing one. In the following, the plasmid containing the new part to be inserted will be referred to as the ''donor'' and the plasmid accepting the new part will be referred to as the ''acceptor''. Composite pars made of parts '''a''' and '''b''' are denoted '''a.b'''.</p><br> | ||
Revision as of 13:39, 14 October 2007

.:: Introduction ::.
Our aim is to engineer a biological model system which exhibits [http://en.wikipedia.org/wiki/Learning learning ability] and can also work according to [http://en.wikipedia.org/wiki/Divergent_evolution the mechanism of divergent evolution], i.e. a system which can alter its behavior according to external stimuli. We are interested in such a system, since learning and adaptation are playing major roles in living organisms and machine learning has numerous applications in engineering - it is therefore a fantastic interface between engineering and biology. Possible applications are as exciting as biological memories or self-adaptative systems.
.:: A Biological Multiple State Automaton ::.
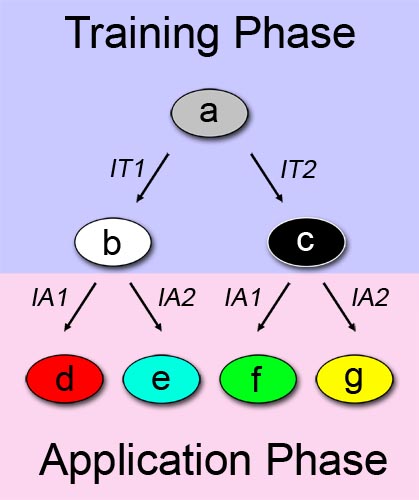
A straightforward approach on how to describe learning behavior and adaptive evolution, can be see in Figure 1. We can separate the process in two phases: a training or learning phase (shown in blue), and an application or "real world" phase (shown in pink), and describe our system as a multiple state automaton. The examined system can alter its state according to a certain input/stimuli that it was exposed to in the first phase. In this binary example, the system changes its state from a to b when it is exposed to a first training-phase-input (IT1). Similarly, the system changes its state from a to c when the other training-phase-input (IT2) is applied.
The principle described above is also valid in the application phase. In the application phase, we have two possibilities for initial system state. Thus, the automaton expands with four more states. Depending on which state the system is at the start of the application phase, and which chemical it is exposed to, it reaches a different final state. State d is reached if the first out of two possible application-phase-inputs (IA1) is applied while the system is at state b. In the end, we can differentiate between the possibilities of the system being trained with one chemical, and being exposed to a different chemical in the application phase. Since we have two different training chemicals, and two different application chemicals, we reach the final number of four states.
In the following, we elaborate on the two phases of our system:
- How can we describe learning ability with this approach?
Define the training-phase-inputs themselves as the information entities to be learned. This implies that after the training phase, the information is permanently stored in the system - a memory has been created. According to its memory, the system will behave differently when it is exposed to a certain stimuli in a later stage. - If one have a look at Figure 1 once more, one can easily spot similarity to family trees or [http://en.wikipedia.org/wiki/Phylogenetic_tree phylogenetic trees]. This raises the concept of divergent evolution (adaptation). There are several differences to the learning model: First of all, the states don't describe a single living entity with changing characteristics but different populations or species. Secondly, the training-phase-inputs and application-phase-inputs are not related to information inputs but rather to events acting on those populations/species. Thirdly, there is no specific training- and application phase but just two phases with different events/stimuli acting on the populations/species.
The following example might show this concept more clearly: Let's say the population of precursor (= ancestor) species a is splitted into two subpopulations (a1 and a2) due to emigration of subpopulation a1 to another geographic region. Application-phase-input 1 (AI1) would then equal "emigration" (a -> a2) and application-phase-input 2 (AI2) would be "no emigration" (a -> a1). The isolated populations then undergo changes as they (1.) become subjected to dissimilar selective pressures or (2.) they independently undergo genetic drift. When the populations come back into contact, they have evolved such that they are reproductively isolated and are no longer capable of exchanging genes [1]. Therefore, subpopulation a1 has evolved into species b, whereas subpopulation a2 has evolved into species c.
This example explains the principle of [http://en.wikipedia.org/wiki/Allopatric_speciation allopatric speciation]. However, our model is not limited to this: Peripatric speciation, parapatric speciation, sympatric speciation or artificial speciation can also be expressed through this model system. Another highly exciting aspect of our model system is, that it can describe Evolution without changing the DNA content over time! How this can be achieved is explained in more detail in the following sections. - (The automaton of Fig. 1 presents similarities to family trees, or [http://en.wikipedia.org/wiki/Phylogenetic_tree phylogenetic trees]. According to different external stimuli, the initial population divides, and evolves into different species. The proposed automaton model can be used to explain the concepts of peripatric speciation, parapatric speciation, sympatric speciation or artificial speciation e.t.c. However, the most imporant fact, and what has stimulated our research in the area, is, to create a biological system that can evolve without changing its DNA content over time).
.:: Detailed System ::.
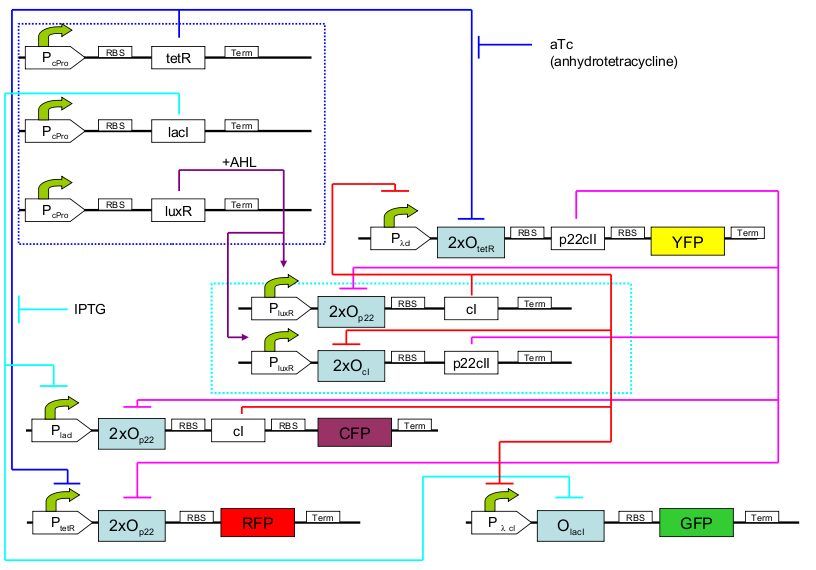
In our project we are constructing an E. coli strain which with the help of an external chemical signal (AHL) is able to remember which of the two chemical substances (aTc and IPTG) it has previously been exposed to. The system architecture is based on a toggle switch consisting of different repressor and activator proteins synthesized from promoters which subject to two different regulations. The full system can be seen in Fig. 2. In the first operation phase (learning), the system is exposed to one of the two chemicals (aTC and IPTG) and AHL is added, causing a steady system behavior. In the second phase (remembering), the chemicals are removed, but AHL allows the system to still maintain its state. Finally, in the final phase (recognition), the system is exposed to any of the two chemicals again. Its response, reported with four fluorescent proteins, differs according not only to which chemical the system is exposed to now, but also to if this chemical is the same that the system has already experienced (learning effect). Therefore, four possible system responses are possible:- now exposed and initially trained with aTc
- now exposed but initially not trained with aTc
- now exposed and initially trained with IPTG
- now exposed and initially not trained with IPTG
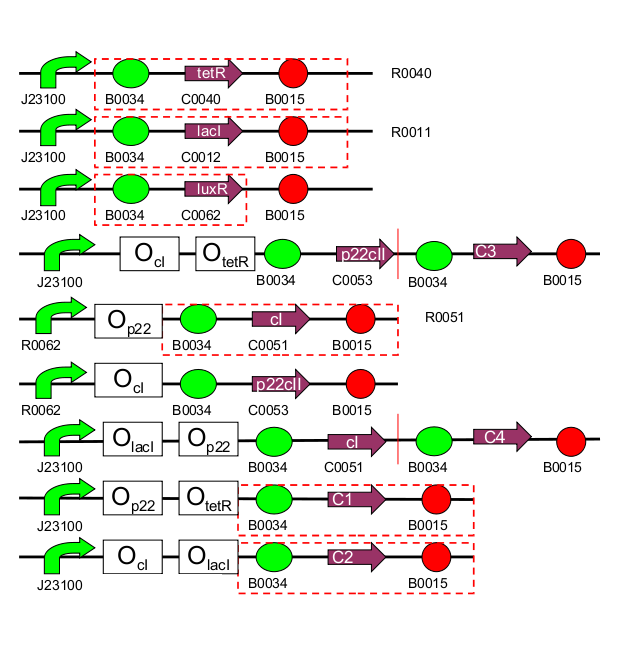
| 1 | [http://partsregistry.org/Part:BBa_I739001 TetR production] (BBa_I739001): constitutive part of system |
|---|---|
| 2 | [http://partsregistry.org/Part:BBa_I739002 LacI production] (BBa_I739002): constitutive part of system |
| 3 | [http://partsregistry.org/Part:BBa_I739003 LuxR production] (BBa_I739003): constitutive part of system |
| 4 | 1st half of p22/YFP production (outer part of system, reporting) |
| 5 | [http://partsregistry.org/Part:BBa_I739005 2nd half of p22/EYFP production] (BBa_I739005): outer part of system, reporting |
| 6 | CI production (inner part of system) |
| 7 | p22 production (inner part of system) |
| 8 | 1st half of CI/CFP production (outer part of system, reporting) |
| 9 | [http://partsregistry.org/Part:BBa_I739009 2nd half of CI/ECFP production] (BBa_I739009): outer part of system, reporting |
| 10 | RFP production (reporting) |
| 11 | GFP production (reporting) |
| 1 | [http://partsregistry.org/Part:BBa_I739101 PoC promoter] (BBa_I739101): Proof of concept |
|---|---|
| 2 | [http://partsregistry.org/Part:BBa_I739102 (cI negative / TetR negative) promoter] (BBa_I739002): inner part of system, no part of the system |
.:: Experiments ::.
.:: References ::.
[1] Standard Assembly Process, http://partsregistry.org/Assembly:Standard_assembly
.:: To Do ::.
- Katerina: 1. Number system parts on both figures for easier reference.
- Katerina: 2. Add more info on all system parts and link to the ones existing in the registry. Write info on the ones that didn't exist in the registry (with detailed info such as addition of bp's as Christian and Sven had done).
- Katerina: 3. Add cloning plan. (Christos: Maybe the details will be at the team note's?)
Martin: I wouldn't put it here, I mean the cloning plan is really nothing special, it's like an auxiliary calculation for a polynom division... Not exciting and everyone could do it... Please correct me if I'm wrong. By the way I wouldn't write so much about the methods with which we cloned the parts in. These are generally known and every biologist knows them. So I think a jury member who knows all the stuff (which he really should) will be bored if he has to work through the whole assembly and plasmid stuff. I would just say, that we use three Plasmids (with names) for insertion of our system into the bacteria and I would also write which part is on which plasmid, but not more. I would more concentrate on the System, how it's working, what parts we've used and so on.... - Christos: 1. The picture with the abstraction - maybe we can put better names on the arrows.
- Christos: 2. We can make figure 1 more specific. Instead of having IT and IA, we can have Chemical 1, Chemical 2, and then again. I think it will be more obvious like that.
- Christos: 3. Nice descriptions Stefan. However, I made the text less, and kept the original idea, since I felt like it was moving away from the purpose, which is a simple introduction and clarification of concepts. I also removed the pictures. I have backups of everything, so, we can put it back if the others disagree.
- Christos: 4. Figure 3 needs to be replaced with the new parts.
- Christos: 5. Are we sure the plasmids that we say are the correct ones? Sven said they were changed, but I didn't really get it.
Martin: Yes, they are the right ones... If not, the shit is hitting the fan - albeit in my opinion it has already hit the fan, but not due to the plasmids more due to Genefart...I can really assure you, that they are right. ;-)