ETHZ/Biology
From 2007.igem.org
Stefan Luzi (Talk | contribs) |
(→.:: System phases ::.) |
||
| Line 119: | Line 119: | ||
| yes | | yes | ||
| no | | no | ||
| - | | | + | | yes |
| yes | | yes | ||
| no | | no | ||
| - | | | + | | yfp |
|- | |- | ||
| Trained with aTc<br>Tested with IPTG | | Trained with aTc<br>Tested with IPTG | ||
| no | | no | ||
| + | | yes | ||
| + | | yes | ||
| yes | | yes | ||
| no | | no | ||
| - | | | + | | gfp |
| - | + | ||
| - | + | ||
|- | |- | ||
| Trained with IPTG<br>Tested with IPTG | | Trained with IPTG<br>Tested with IPTG | ||
| no | | no | ||
| yes | | yes | ||
| - | | | + | | yes |
| no | | no | ||
| yes | | yes | ||
| - | | | + | | cfp |
|- | |- | ||
| Trained with IPTG<br>Tested with aTc | | Trained with IPTG<br>Tested with aTc | ||
| yes | | yes | ||
| no | | no | ||
| + | | yes | ||
| no | | no | ||
| - | | | + | | yes |
| - | + | ||
| rfp | | rfp | ||
|- | |- | ||
Revision as of 12:25, 19 October 2007

In this page, you can find an analysis of the function of our system and its relation to epigenetics, its biological design and a list of the parts that it consists of. Are you interested in constructing educatETH E.coli in your lab? Then under In the Lab, you can find the ingredients and equipment we used, the electronic version of our lab notebook and a presentation of all the difficulties that we encountered. If you are also interested on how educatETH E.coli was simulated outside the lab, please visit the Engineering Perspective.
Contents |
.:: Introduction ::.
educatETH E.coli is a system which can distinguish between aTc and IPTG based on a previous training phase conducted with the same chemicals and the help of AHL. It composes of three subsystems: the subsystem of constitutively produced proteins, the learning subsystem and the reporting subsystem. The constitutively produced proteins (lacI, TetR and LuxR) control the learning subsystem. At the core of the latter there exists a modified version of the toggle switch found in [1] with two operator sites, so that it only changes its state when both one of the two chemicals (aTc/IPTG) and AHL are present. As AHL is only present during the training phase, the toggle maintains its state during testing, and thus can “memorize”. In the reporting subsystem, four reporters allow supervision of both the chemical the system was trained with and of if the system recognizes the chemical it is being exposed to in the testing phase as one it has been trained with or not.
.:: The whole system ::.
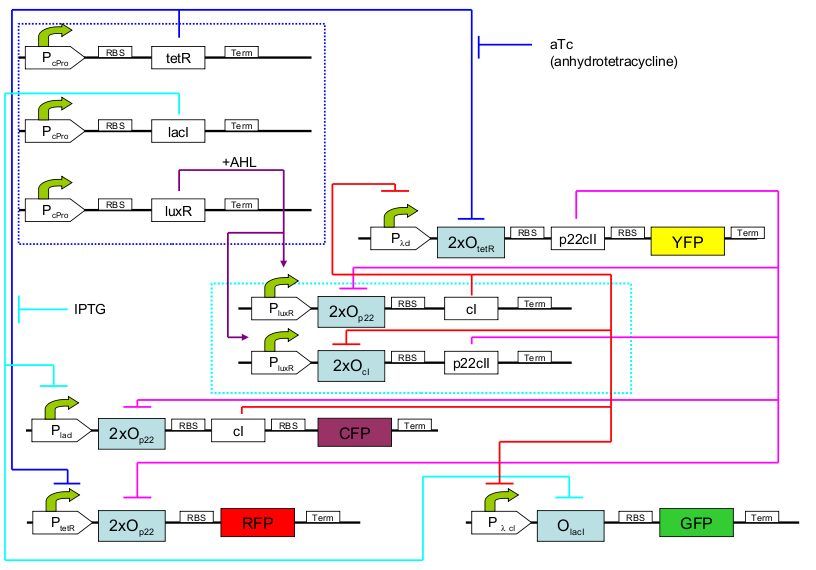
The biological design of educatETH E.coli is presented on
. In the following, we will clarify the function of all depicted components. (Are you interested in how the complex system of Fig. 1 was modelled? Then visit the Engineering Perspective!).:: Constitutive subsystem ::.
The constitutively produced proteins of the system are LacI, TetR and LuxR. The LuxR part has a special function: when AHL is present, it forms a LuxR-AHL complex which acts on the learning subsystem (more on this later). At the moment, we will consider that AHL is absent and therefore LuxR cannot act on any subsystems. The TetR and lacI parts behave similarly: more specifically, the tetR protein in the absence of aTc inhibits the production of p22cII and LacI in the absence of IPTG inhibits the production of cI. When aTc is present, however, the p22cII production is no longer inhibited (and thus aTc is produced). Respectively, cI is produced when IPTG is present.
.:: Learning subsystem ::.
The learning subsystem is a toggle switch with two operator sites. The upper part of the toggle (cI production) has operator sites for the LuxR-AHL complex and p22cII (which has been in turn induced by aTc). The LuxR-AHL complex induces the cI production, whereas p22cII inhibits it. The lower part of the toggle (p22cII production) has operator sites for the LuxR-AHL comple and cI (which has been induced by IPTG). Similarly with the upper part, the LuxR-AHL complex induces the p22cII production and cI inhibits it. Therefore, the switch always requires the presence of the LuxR-AHL complex in order for it to operate. Its state depends on the presence of p22cII and cI into the system, which in curse was caused through the exposure of the system to aTc and IPTG.
.:: Reporting subsystem ::.
There are four reporters in the system. CFP and YFP are active during the training phase of the system and show which chemical the system is exposed to during training, whereas GFP and RFP are active during the testing phase and show if the system is exposed to the same chemical as in training or not. More specifically, the YFP protein production is regulated with help of two operator sites controlled by cI and aTc. cI inhibits YFP production and aTc induces it. Therefore, YFP is produced when the system is exposed to aTc. In a similar manner, the CFP production is produced when the system is exposed to IPTG. The GFP production is regulated with help of two operator sites controlled by lacI and .
.:: System phases ::.
The system operation is divided into two main phases: the training phase and the testing phase. The training phase itself is also subdivided into two phases: seeing and memorizing. During seeing, the system is first exposed to one of the two chemicals it is designed to recognize (aTc and IPTG). AHL is then added and the system’s internal toggle switch reaches a steady state. During memorizing, the chemical used during seeing is removed and only AHL is retained. This maintains the toggle switch to its acquired steady state, which is reported with YFP (if aTc was seen) or CFP (if IPTG was seen). During the testing phase, the system is exposed to any of the two chemicals (aTc or IPTG), but not to AHL. By comparing its toggle switch state with the effect of the newly introduced chemical, the system shows a different response if it has previously been exposed to this chemical (GFP) or not (RFP).The following table presents all possible paths that may be taken by the system during all phases of operation according to the external stimuli.
| aTc | IPTG | AHL | p22cII | cI | Reporting | |
|---|---|---|---|---|---|---|
| Start | no | no | no | no | no | no |
| Learning | ||||||
| Seeing | ||||||
| Trained with aTc | yes | no | no | yes | no | yfp |
| Trained with IPTG | no | yes | no | no | yes | cfp |
| Memorizing | ||||||
| Trained with aTc | yes | no | yes | yes | no | yfp |
| Trained with IPTG | no | yes | yes | no | yes | cfp |
| Testing | ||||||
| Trained with aTc Tested with aTc | yes | no | yes | yes | no | yfp |
| Trained with aTc Tested with IPTG | no | yes | yes | yes | no | gfp |
| Trained with IPTG Tested with IPTG | no | yes | yes | no | yes | cfp |
| Trained with IPTG Tested with aTc | yes | no | yes | no | yes | rfp |
.:: Further thoughts on the system phases ::.
TODO: put Stefan's new text here. Text should be smaller.
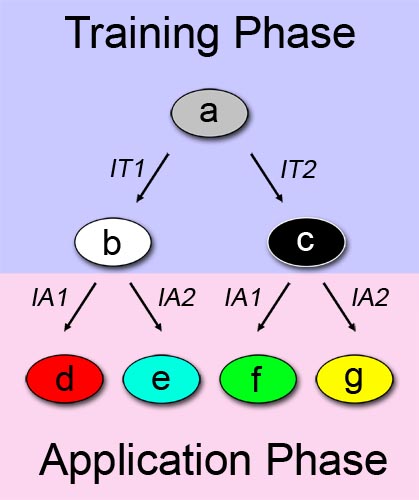
A straightforward approach on how to describe learning behavior and adaptive evolution, can be see in Figure 1. We can separate the process in two phases: a training or learning phase (shown in blue), and an application or "real world" phase (shown in pink), and describe our system as a multiple state automaton. The examined system can alter its state according to a certain input/stimuli that it was exposed to in the first phase. In this binary example, the system changes its state from a to b when it is exposed to a first training-phase-input (IT1). Similarly, the system changes its state from a to c when the other training-phase-input (IT2) is applied.
The principle described above is also valid in the application phase. In the application phase, we have two possibilities for initial system state. Thus, the automaton expands with four more states. Depending on which state the system is at the start of the application phase, and which chemical it is exposed to, it reaches a different final state. State d is reached if the first out of two possible application-phase-inputs (IA1) is applied while the system is at state b. In the end, we can differentiate between the possibilities of the system being trained with one chemical, and being exposed to a different chemical in the application phase. Since we have two different training chemicals, and two different application chemicals, we reach the final number of four states.
In the following, we elaborate on the two phases of our system:
- How can we describe learning ability with this approach?
Define the training-phase-inputs themselves as the information entities to be learned. This implies that after the training phase, the information is permanently stored in the system - a memory has been created. According to its memory, the system will behave differently when it is exposed to a certain stimuli in a later stage. - If one have a look at Figure 1 once more, one can easily spot similarity to family trees or [http://en.wikipedia.org/wiki/Phylogenetic_tree phylogenetic trees]. This raises the concept of divergent evolution (adaptation). There are several differences to the learning model: First of all, the states don't describe a single living entity with changing characteristics but different populations or species. Secondly, the training-phase-inputs and application-phase-inputs are not related to information inputs but rather to events acting on those populations/species. Thirdly, there is no specific training- and application phase but just two phases with different events/stimuli acting on the populations/species.
The following example might show this concept more clearly: Let's say the population of precursor (= ancestor) species a is splitted into two subpopulations (a1 and a2) due to emigration of subpopulation a1 to another geographic region. Application-phase-input 1 (AI1) would then equal "emigration" (a -> a2) and application-phase-input 2 (AI2) would be "no emigration" (a -> a1). The isolated populations then undergo changes as they (1.) become subjected to dissimilar selective pressures or (2.) they independently undergo genetic drift. When the populations come back into contact, they have evolved such that they are reproductively isolated and are no longer capable of exchanging genes [1]. Therefore, subpopulation a1 has evolved into species b, whereas subpopulation a2 has evolved into species c.
This example explains the principle of [http://en.wikipedia.org/wiki/Allopatric_speciation allopatric speciation]. However, our model is not limited to this: Peripatric speciation, parapatric speciation, sympatric speciation or artificial speciation can also be expressed through this model system. Another highly exciting aspect of our model system is, that it can describe Evolution without changing the DNA content over time! How this can be achieved is explained in more detail in the following sections. - (The automaton of Fig. 1 presents similarities to family trees, or [http://en.wikipedia.org/wiki/Phylogenetic_tree phylogenetic trees]. According to different external stimuli, the initial population divides, and evolves into different species. The proposed automaton model can be used to explain the concepts of peripatric speciation, parapatric speciation, sympatric speciation or artificial speciation e.t.c. However, the most imporant fact, and what has stimulated our research in the area, is, to create a biological system that can evolve without changing its DNA content over time).
.:: System parts ::.
educatETH E.coli consists of 11 parts which may be synthesized independently. Please note that four of them (4,5 and 8,9) form together two functional system units. They have been separated to ensure comparable part lengths and thus enable easier introduction into plasmids.
The system consists of 11 parts that can be synthesized independently (want to know how this is done in the lab? Then visit our In the Lab page!)
| 1 | TetR production | [http://partsregistry.org/Part:BBa_I739001 BBa_I739001] | constitutive subsystem | |
|---|---|---|---|---|
| 2 | LacI production | [http://partsregistry.org/Part:BBa_I739002 BBa_I739002] | constitutive subsystem | |
| 3 | LuxR production | [http://partsregistry.org/Part:BBa_I739003 BBa_I739003] | constitutive subsystem | |
| 4 | 1st half of P22 cII / EYFP production | [http://partsregistry.org/Part:BBa_I739004 BBa_I739004] | reporting subsystem | |
| 5 | 2nd half of P22 cII / EYFP production | [http://partsregistry.org/Part:BBa_I739005 BBa_I739005] | reporting subsystem | |
| 6 | cI production | [http://partsregistry.org/Part:BBa_I739006 BBa_I739006] | learning subsystem | |
| 7 | P22 cII production | [http://partsregistry.org/Part:BBa_I739007 BBa_I739007] | learning subsystem | |
| 8 | 1st half of cI / ECFP production | [http://partsregistry.org/Part:BBa_I739008 BBa_I739008] | reporting subsystem | |
| 9 | 2nd half of cI / ECFP production | [http://partsregistry.org/Part:BBa_I739009 BBa_I739009] | reporting subsystem | |
| 10 | RFP production | [http://partsregistry.org/Part:BBa_I739010 BBa_I739010] | reporting subsystem | |
| 11 | GFP production | [http://partsregistry.org/Part:BBa_I739011 BBa_I739011] | reporting subsystem |
| 1+2+3 | tetR + lacI + luxR production | [http://partsregistry.org/Part:BBa_I739013 BBa_I739013] | constitutive subsystem | |
|---|---|---|---|---|
| 4+5 | P22 cII + EYFP production | [http://partsregistry.org/Part:BBa_I739015 BBa_I739015] | reporting subsystem | |
| 8+9 | cI + ECFP production | [http://partsregistry.org/Part:BBa_I739016 BBa_I739016] | reporting subsystem | |
| (4+5)+(8+9) | (P22 cII + EYFP) + (cI + ECFP) production | [http://partsregistry.org/Part:BBa_I739017 BBa_I739017] | reporting subsystem | |
| 6+7 | cI + P22 cII production | [http://partsregistry.org/Part:BBa_I739018 BBa_I739018] | learning subsystem | |
| 10+11 | RFP + GFP production | [http://partsregistry.org/Part:BBa_I739019 BBa_I739019] | reporting subsystem | |
| (6+7)+(10+11) | (cI + P22 cII) + (RFP + GFP) production | [http://partsregistry.org/Part:BBa_I739020 BBa_I739020] | learning/reporting subsystem |
| 1' | cI negative / tetR negative promoter | [http://partsregistry.org/Part:BBa_I739102 BBa_I739102] | reporting subsystem | |
|---|---|---|---|---|
| 2' | lacI negative / P22 cII negative promoter | [http://partsregistry.org/Part:BBa_I739103 BBa_I739103] | reporting subsystem | |
| 3' | luxR/HSL positive / P22 cII negative promoter | [http://partsregistry.org/Part:BBa_I739104 BBa_I739104] | learning subsystem | |
| 4' | luxR/HSL positive / cI negative promoter | [http://partsregistry.org/Part:BBa_I739105 BBa_I739105] | learning subsystem | |
| 5' | tetR negative / P22 cII negative promoter | [http://partsregistry.org/Part:BBa_I739106 BBa_I739106] | reporting subsystem | |
| 6' | cI negative / lacI negative promoter | [http://partsregistry.org/Part:BBa_I739107 BBa_I739107] | reporting subsystem |
| 1" | PoC promoter | [http://partsregistry.org/Part:BBa_I739101 BBa_I739101] | proof of concept, no part of the system | |
|---|---|---|---|---|
| 2" | PoC intermediate | [http://partsregistry.org/Part:BBa_I739014 BBa_I739014] | proof of concept, no part of the system | |
| 3" | PoC composite | [http://partsregistry.org/Part:BBa_I739021 BBa_I739021] | proof of concept, no part of the system |
.:: References ::.
[1] Standard Assembly Process, http://partsregistry.org/Assembly:Standard_assembly
.:: To Do ::.
.:: New ::.
- Update and correct parts in parts list. Write better in a table
- Update and correct full system scheme
- Update graph scheme (made by Stefan) using aTc, IPTG instead of it1,2 and ia1,2
- Which reporters are active when? I think CFP and YFP are not active only during training. Change text if needed.
- Proposed terminology: seeing, memorizing
- What are GFP, RFP controlled by? Is the full system scheme correct there?
- What are the “double promoters” mentioned?
- Check my terminology (operator sites etc)
- Put Stefan's updated part on epigenetics
- How was Sven’s standard notation on how to write differently proteins, dna, rna?
- Fill in table completely, make it more reading-friendly
- Make "In the lab page", replace links.
- Put image with 11 system parts (updated one, created by katerina)
.:: Old::.
- Katerina: 1. Number system parts on both figures for easier reference.
- Katerina: 2. Add more info on all system parts and link to the ones existing in the registry. Write info on the ones that didn't exist in the registry (with detailed info such as addition of bp's as Christian and Sven had done).
- Katerina: 3. Add cloning plan. (Christos: Maybe the details will be at the team note's?)
Martin: I wouldn't put it here, I mean the cloning plan is really nothing special, it's like an auxiliary calculation for a polynom division... Not exciting and everyone could do it... Please correct me if I'm wrong. By the way I wouldn't write so much about the methods with which we cloned the parts in. These are generally known and every biologist knows them. So I think a jury member who knows all the stuff (which he really should) will be bored if he has to work through the whole assembly and plasmid stuff. I would just say, that we use three Plasmids (with names) for insertion of our system into the bacteria and I would also write which part is on which plasmid, but not more. I would more concentrate on the System, how it's working, what parts we've used and so on.... - Christos: 1. The picture with the abstraction - maybe we can put better names on the arrows.
- Christos: 2. We can make figure 1 more specific. Instead of having IT and IA, we can have Chemical 1, Chemical 2, and then again. I think it will be more obvious like that.
- Christos: 3. Nice descriptions Stefan. However, I made the text less, and kept the original idea, since I felt like it was moving away from the purpose, which is a simple introduction and clarification of concepts. I also removed the pictures. I have backups of everything, so, we can put it back if the others disagree.
- Christos: 4. Figure 3 needs to be replaced with the new parts.
- Christos: 5. Are we sure the plasmids that we say are the correct ones? Sven said they were changed, but I didn't really get it.
Martin: Yes, they are the right ones... If not, the shit is hitting the fan - albeit in my opinion it has already hit the fan, but not due to the plasmids more due to Genefart...I can really assure you, that they are right. ;-)