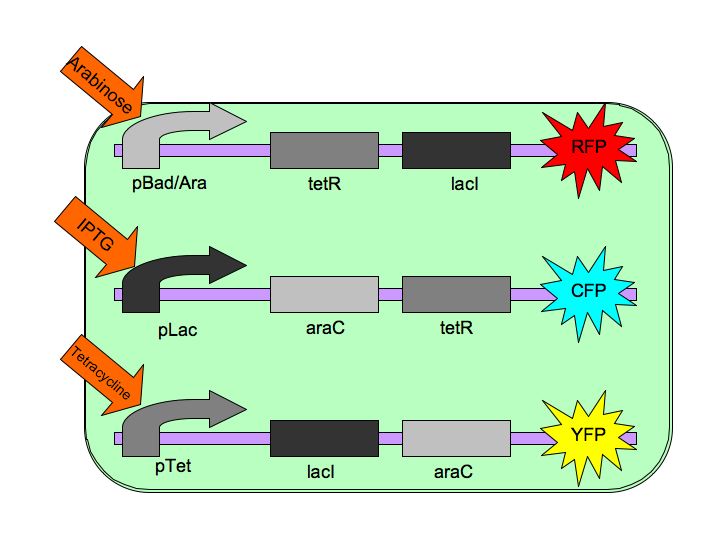
Tristable
From 2007.igem.org
Norrishung (Talk | contribs) |
Norrishung (Talk | contribs) |
||
| Line 67: | Line 67: | ||
<!-- PLACE CONTENT IN HERE--> | <!-- PLACE CONTENT IN HERE--> | ||
| - | |||
| - | |||
| Line 81: | Line 79: | ||
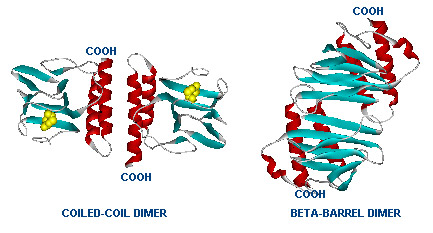
The gene AraC one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html] [[Image:Two_Dimers_of_AraC.jpg|thumb|left|Dimer structure with arabinose on the left (yellow)]] | The gene AraC one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html] [[Image:Two_Dimers_of_AraC.jpg|thumb|left|Dimer structure with arabinose on the left (yellow)]] | ||
[[Image:AraC_Promoters.gif|left|thumb|The left image shows the araC dimer repressing transcription, while the right conformation enables transcription]]The protein forms a dimer in with and without arabinose but the structural change activates or represses the pBAD ([http://en.wikipedia.org/wiki/Bcl-2-associated_death_promoter Bcl-2-associated death promoter], an apoptotic regulator in humans). | [[Image:AraC_Promoters.gif|left|thumb|The left image shows the araC dimer repressing transcription, while the right conformation enables transcription]]The protein forms a dimer in with and without arabinose but the structural change activates or represses the pBAD ([http://en.wikipedia.org/wiki/Bcl-2-associated_death_promoter Bcl-2-associated death promoter], an apoptotic regulator in humans). | ||
| - | |||
| Line 88: | Line 85: | ||
====LacI==== | ====LacI==== | ||
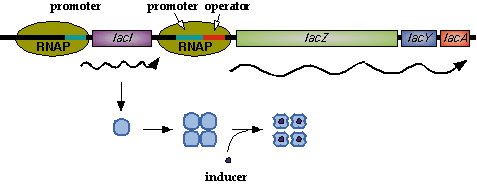
| - | In nature, LacI represses pLac which promotes LacYZA genes that metabolize lactose, thus LacI represses pLac except in the presence of lactose (or lactose mimics, eg IPTG). [[Image: | + | In nature, LacI represses pLac which promotes LacYZA genes that metabolize lactose, thus LacI represses pLac except in the presence of lactose (or lactose mimics, eg IPTG). [[Image:LacI_repressor.gif|thumb|left|Image[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html]. LacI forms a tetramer and represses pLac. However, an inducer, such as IPTG, causes a conformation change that removes LacI from the operator site.]] Lactose causes a conformational change which inhibits LacI from binding to the operator site of pLac. Four LacI proteins form a tetramer to inhibit pLac and four inducer molecules are required to cause the full conformational change in the inhibitor.[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html] |
| + | |||
| + | |||
| + | |||
| + | |||
| + | ====TetR==== | ||
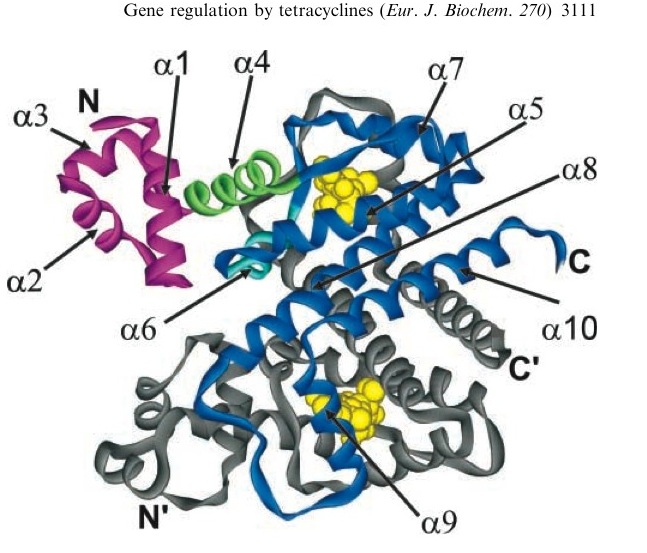
| + | TetR represses the constitutive promoter pTet. In the presence of tetracycline, an antibiotic, a conformational change in TetR inhibits the protein from binding to the operator region. In nature, pTet promotes TetR and TetA. The latter which acts to pump tetracycline out of the cell, thus the pump is only activated in the presence of Tetracycline.[http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation] | ||
| + | The TetR, as it turns out is a very tight repressor and a range of 0 to 1 ug/ml has been shown to cause a 5 order of magnitude change in luciferase production.[http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1319065&query_hl=1&itool=pubmed_docsum] | ||
| + | [[Image:Tc_bound_to_TetR.jpg|thumb|left|A tetracycline molecule binds to each of the two TetR monomers to form a dimer]] | ||
| + | Tetracycline is highly diffusable through cell membrane (permeation coeficient or 5.6±1.9 * 10^-9 cm/s or half equilibrium time = 35 ± 15 min) and TetR shows a very high affinity for the molecule. The binding constant of TetR to [tc-Mg+] is Ka ~ 10^9 M^-1. When bound to tc, TetR has a low binding level to DNA of 10^5 M^-1. [http://content.febsjournal.org/cgi/content/abstract/270/15/3109] | ||
| + | |||
| + | |||
| + | |||
| + | ===Modeling=== | ||
| + | Brown iGEM 2006 Matlab model code [[Media:tristable2006.txt]] | ||
| + | |||
| + | [http://parts2.mit.edu/wiki/index.php/Table_of_preliminary_model_constants Initial Table of Constants] | ||
| + | |||
| + | [http://parts2.mit.edu/wiki/index.php/Derivation_of_the_Model_Equations Derivation of Model Equations] | ||
| + | A simpler model based on the bistable paper was developed that takes the relative transcription/translation rates into account [[Media:tristable2007.txt]]. | ||
Revision as of 21:27, 20 October 2007

| |||||||||||||||||