USTC/Repressor Evolution on Plates
From 2007.igem.org
Xiaofeng Su (Talk | contribs) |
Xiaofeng Su (Talk | contribs) m |
||
| Line 75: | Line 75: | ||
== Results == | == Results == | ||
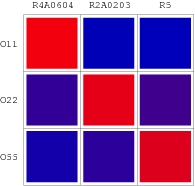
| - | The consequent data of reporter's expression is, by formula, converted to the Repression Value(R.V.) representing the repression intensity of each repressor-promoter pair. The sets of R.V. is depicted on the scheme below which have been | + | The consequent data of reporter's expression is, by formula, converted to the Repression Value(R.V.) representing the repression intensity of each repressor-promoter pair. The sets of R.V. is depicted on the scheme below which have been transformed to corresponding Repression Matrix-more visual that the coordination sheme. And the Orthogonal Repression Matrix birthing from Repression Matrix can be used to acquire specific repressor-promoter pairs with high specificity. |
=== Repression Scheme === | === Repression Scheme === | ||
Revision as of 05:47, 23 October 2007
Contents |
Wires in Bio-logic Circuit
In electronic circuit, metal conductor such as copper is used as wires widely. But in a cell, most of the things are diffusive, so it is a difficult problem to limit a signal in a specific signal channel. High-specific regulator-operator pairs can be used as "copper wires" in bio-logic circuit. It means, repressor or activator transmit a particular signal from the upstream output port to the downstream input port, without interference between each other, like carrier wave in FM radio.
Why to Design Artificial High-Specific Repressor-Operator Pairs
We have been attempting to construct several artificial high-specific repressor-operator pairs to serve as the connecting wires of our system, based on the knowledge of LacR and its binding site, and by means of directed evolution and computational protein design. We decide that they should be newly designed for several reasons,
- The number of natural regulators well studied is limited.
- Natural regulators do have some disadvantages.
- For example, it is well known that there are dozens of downstream regulatory sites of CRP in E.Coli, and if we abuse the CRP, some other natural pathways in the host bacterial will probably be interrupted.
- There may be several unknown sites to be bound with the selected native activators, and we might get unexpected results in such situations.
- This artificial repressor family hold the same property for our cis-acting logic promoters, such as dimerization ad tetramerization.
Start: Lac repressor
[http://en.wikipedia.org/wiki/Lac_repressor From Wikipedia]:
The lac repressor is a DNA-binding protein which inhibits the expression of genes coding for proteins involved in the metabolism of lactose in bacteria. It is active in the absence of lactose, ensuring that the bacterium only invests energy in the production of machinery necessary for the uptake and metabolism of lactose when lactose is present. When lactose becomes available, it is converted into allolactose, which inhibits the lac repressor's DNA binding ability.
The lac repressor protein has three distinct regions:
- a core region (which binds allolactose)
- a tetramerization region (which joins four monomers in an alpha-helix bundle)
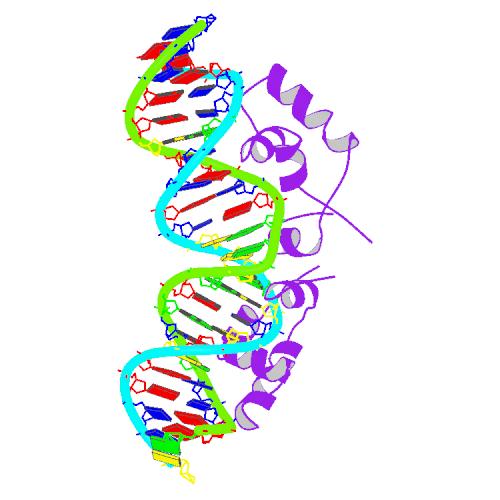
- a DNA-binding region (in which two LacI proteins bind a single operator site)
The lac repressor occurs as a tetramer (four identical subunits bound together). This can be viewed as two dimers, with each dimer being able to bind to a single lac operator. The two subunits each bind to a slightly separated (major groove) region of the operator. The promoter is slightly covered by the lac repressor so RNAP cannot bind to and transcribe the operon.
The DNA binding region consists of a helix-turn-helix structural motif.
Selection of operator sequence
By means of bioinformatics we can select a DNA sequence that has never appeared in the genome of E.Coli, and let the regulator bind to the sequence with quite high specificity,. Therefore, we will not have to worry about the regulator’s interrupting the normal functioning of the host genome (A tiny exception: the host strain should not contain lacI and lac promoter, such as [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 Top10]).
O11 aattgtgagcgctcacaatt O22 aattgtaagcgcttacaatt O33 aattgtaaacgtttacaatt O44 aattgtgaacgttcacaatt O55 aattttgagcgctcaaaatt O66 aattatgagcgctcataatt O77 gacgactgtatacagtcgtc
Directed Repressor Evolution
Targeted Mutagenesis
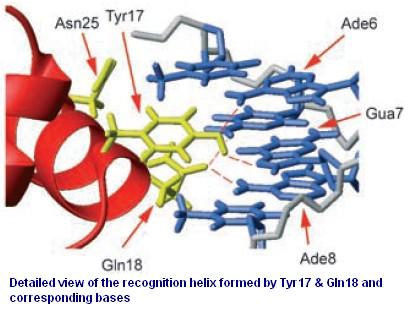
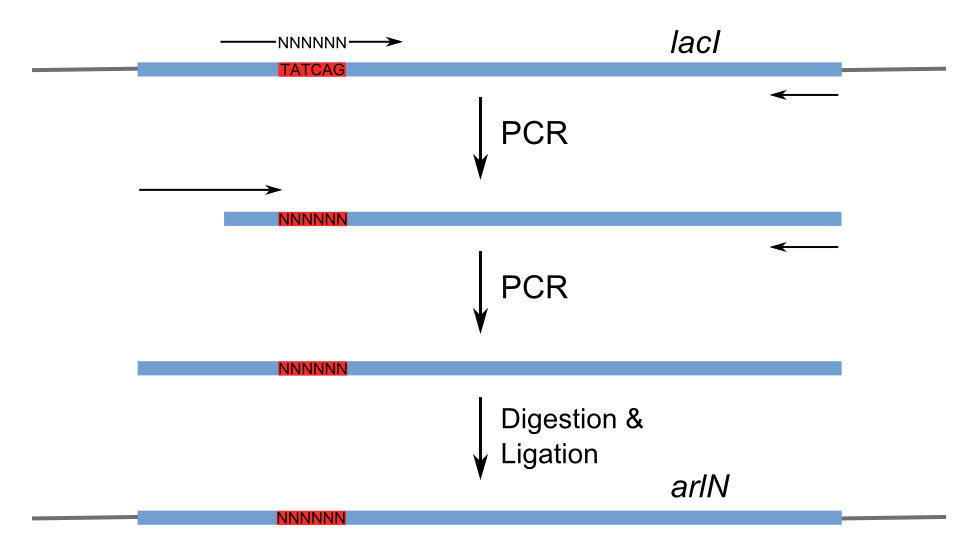
Figure 3 shows the recognition region of which the amino acid residues we will try to modify. With saturation mutagenesis, it is possible to create a library of mutatants containing all possible mutations at these positions. For the targeted sites are near to the beginning of the lacI gene, but beyond the range of one primer region, two steps of PCR are carried out to generate the random repressor family like Figure 4. Then the PCR production is purified and digested, loaded into the repressor-production plasmid to take the repression assay.
Repression Assay
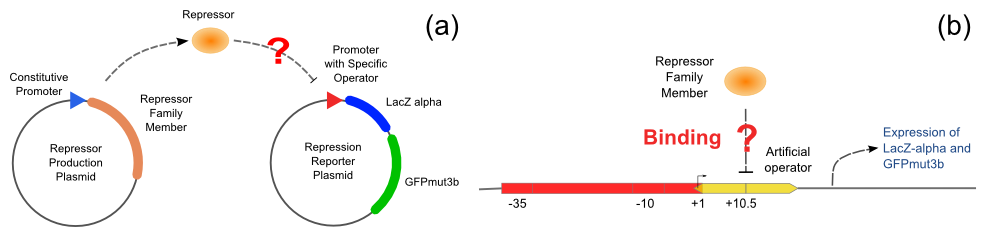
With the help of pUC-repressor and pZS*-reporter, the two plasmids shown in Figure 5, we will be able to accomplish the repression assay.
The first plasmid is in charge of repressor production, that is, expressing the repressor constitutively. The second plasmid is composed of two reporter genes, lacZ (alpha fragment) and gfp-AAV, respectively, and an upstream promoter that reflects the repression effect in the form of binding intensity. Once the repression exists, the promoter will lose its activity. Consequently, neither of the downstream lacZ or GFP-AAV reporter gene will work. By reading the blue/white colonies on a plate with naked eyes and the fluorescence intensity of GFP with a fluorescence microscope, we can finally get to know the repression effect, and can ratiocinate from it the binding affinity of the repressor-operator pair.
Screening
First, pUC-repressor plasmid containing random repressor family members are transformed into Top10/pZS*-PO_target-lacZa. White colonies, in which repressors can repress the target promoter, are select from Blue/White Screening on top agar Luria-Bertani broth. The repression should be re-tested to eliminate the false positive samples by X-gal assay and PCR checking. The survivals will be quantificationally measured using ONPG assay or fluorescent assay.

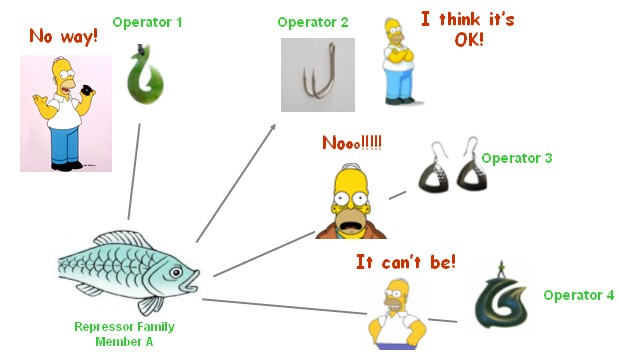
Quantitive Assay(Cross Repression Test)
We selected 7 repressor-promoter pair candidates from Blue/White Screening results above for quantitive assay of specificity and affinity. In addition, 2 existed represor-promoter pairs are added to this work as new candidates. Then, each repressor-expression plasmid is transform to each Top10 competent cell with specific target promoters. Eventually, each expression quantity of LacZ alpha or GFP is measured by ONPG assay(LacZ) or fluorescent assay(GFP).
Results
The consequent data of reporter's expression is, by formula, converted to the Repression Value(R.V.) representing the repression intensity of each repressor-promoter pair. The sets of R.V. is depicted on the scheme below which have been transformed to corresponding Repression Matrix-more visual that the coordination sheme. And the Orthogonal Repression Matrix birthing from Repression Matrix can be used to acquire specific repressor-promoter pairs with high specificity.
Repression Scheme
Repression Matrix
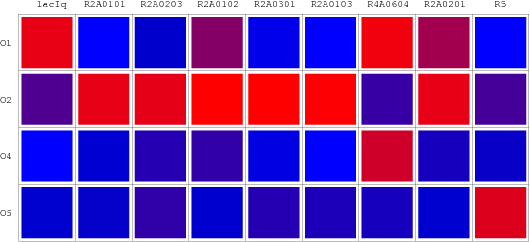
Orthologal Repression Matrix