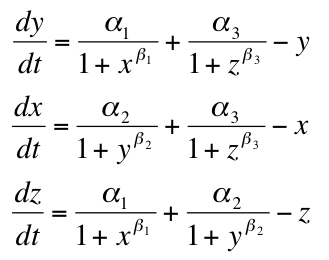Tristable
From 2007.igem.org
| Line 86: | Line 86: | ||
| - | + | ==AraC/BAD== | |
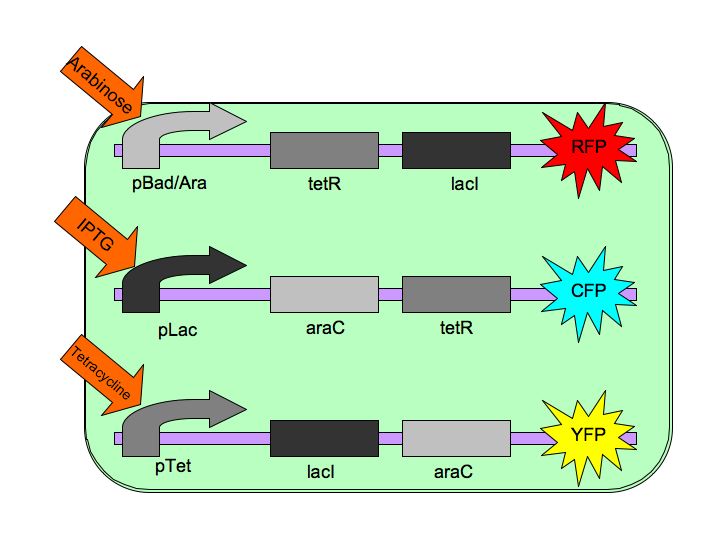
The gene AraC, one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html] | The gene AraC, one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html] | ||
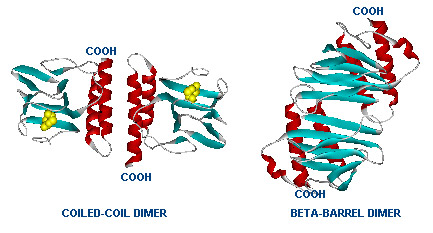
[[Image:Two_Dimers_of_AraC.jpg|thumb|left|Dimer structure with arabinose on the left (yellow)]] | [[Image:Two_Dimers_of_AraC.jpg|thumb|left|Dimer structure with arabinose on the left (yellow)]] | ||
| Line 94: | Line 94: | ||
| - | + | ==LacI== | |
| - | + | ||
| - | + | ||
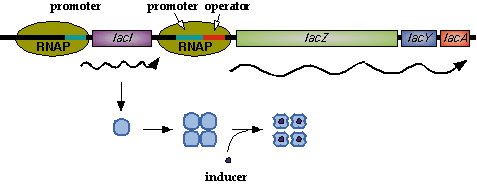
In nature, LacI represses pLac which promotes the LacYZA genes that metabolize lactose. Thus LacI represses pLac except in the presence of lactose (or lactose mimics, eg IPTG). [[Image:LacI_repressor.gif|thumb|left|Image[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html]. LacI forms a tetramer and represses pLac. However, an inducer, such as IPTG, causes a conformation change that removes LacI from the operator site.]] Lactose causes a conformational change which inhibits LacI from binding to the operator site of pLac. Four LacI proteins form a tetramer to inhibit pLac and four inducer molecules are required to cause the full conformational change in the repressor.[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html] | In nature, LacI represses pLac which promotes the LacYZA genes that metabolize lactose. Thus LacI represses pLac except in the presence of lactose (or lactose mimics, eg IPTG). [[Image:LacI_repressor.gif|thumb|left|Image[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html]. LacI forms a tetramer and represses pLac. However, an inducer, such as IPTG, causes a conformation change that removes LacI from the operator site.]] Lactose causes a conformational change which inhibits LacI from binding to the operator site of pLac. Four LacI proteins form a tetramer to inhibit pLac and four inducer molecules are required to cause the full conformational change in the repressor.[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html] | ||
| Line 106: | Line 104: | ||
| - | + | ==TetR== | |
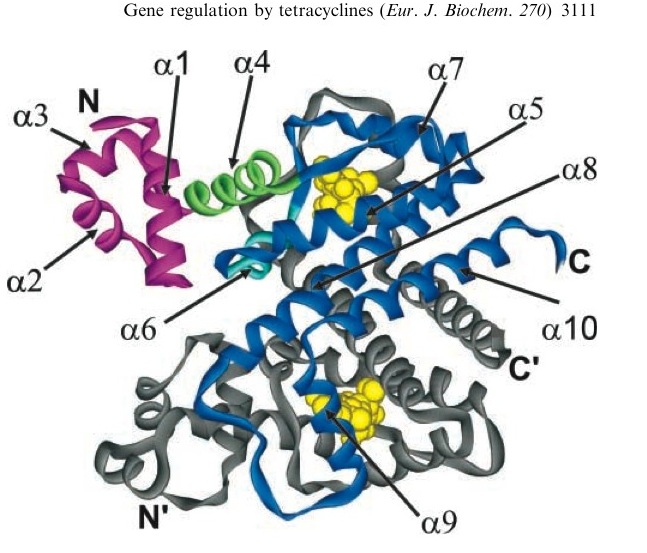
TetR represses the constitutive promoter pTet. In the presence of tetracycline, an antibiotic, a conformational change in TetR inhibits the protein from binding to the operator region. In nature, pTet promotes TetR and TetA. The latter of which acts to pump tetracycline out of the cell, thus the pump is only activated in the presence of Tetracycline.[http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation] | TetR represses the constitutive promoter pTet. In the presence of tetracycline, an antibiotic, a conformational change in TetR inhibits the protein from binding to the operator region. In nature, pTet promotes TetR and TetA. The latter of which acts to pump tetracycline out of the cell, thus the pump is only activated in the presence of Tetracycline.[http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation] | ||
The TetR, as it turns out is a very tight repressor and a range of 0 to 1 ug/ml has been shown to cause a 5 order of magnitude change in luciferase production.[http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1319065&query_hl=1&itool=pubmed_docsum] | The TetR, as it turns out is a very tight repressor and a range of 0 to 1 ug/ml has been shown to cause a 5 order of magnitude change in luciferase production.[http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1319065&query_hl=1&itool=pubmed_docsum] | ||
| Line 128: | Line 126: | ||
| - | + | =Modeling= | |
The first draft of the code started last year: | The first draft of the code started last year: | ||
| Line 138: | Line 136: | ||
A simpler model based on the bistable paper was developed that takes the combined, relative transcription/translation rates into account [[Media:tristable2007.txt]]. | A simpler model based on the bistable paper was developed that takes the combined, relative transcription/translation rates into account [[Media:tristable2007.txt]]. | ||
| - | + | ==Parameters in the Model== | |
| - | + | ===Repressor Production Rate=== | |
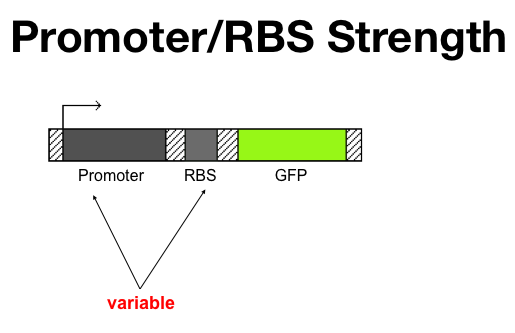
Repressor production rate is determined by two factors: | Repressor production rate is determined by two factors: | ||
#Promoter Strength (Transcription) | #Promoter Strength (Transcription) | ||
| Line 148: | Line 146: | ||
The promoter strength cannot be easily changed because this would require mutations in the promoter or a different promoter/repressor combo. However, RBSs have been well characterized and the alpha parameter can be modulated by inserting different strength RBSs as determined by the model and testing. | The promoter strength cannot be easily changed because this would require mutations in the promoter or a different promoter/repressor combo. However, RBSs have been well characterized and the alpha parameter can be modulated by inserting different strength RBSs as determined by the model and testing. | ||
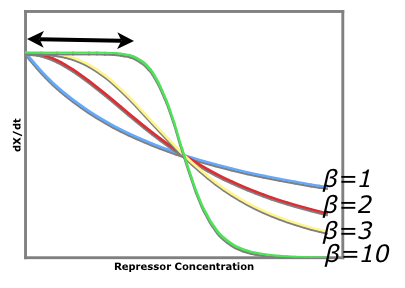
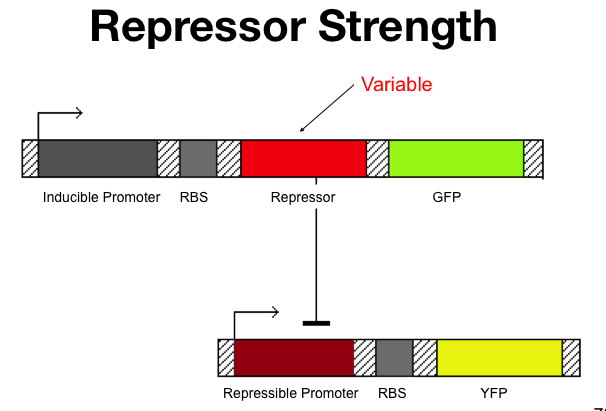
| - | + | ===Repressor Strength=== | |
The strength of the repressors is determined by: | The strength of the repressors is determined by: | ||
#The repressor concentration, [Repressor]. | #The repressor concentration, [Repressor]. | ||
| Line 157: | Line 155: | ||
The Beta value is characteristic of each repressor and represents a constant that cannot be changed. | The Beta value is characteristic of each repressor and represents a constant that cannot be changed. | ||
| - | + | ===Degradation=== | |
The degradation in the model is denoted by the term "-repressor," where the degradation constant is unity because the rates are relative, rather than absolute. The next iteration of the model will include degradation constants. Currently in the registry all of the repressors have strong degradation tags. Jason Kelly made a comment on one repressor and said he was suspicious of how strong of a repression would be able to be obtained from the repessors. We need strong repression for our system, so we may need to remove the tag our system to function. | The degradation in the model is denoted by the term "-repressor," where the degradation constant is unity because the rates are relative, rather than absolute. The next iteration of the model will include degradation constants. Currently in the registry all of the repressors have strong degradation tags. Jason Kelly made a comment on one repressor and said he was suspicious of how strong of a repression would be able to be obtained from the repessors. We need strong repression for our system, so we may need to remove the tag our system to function. | ||
| - | + | ==The Model== | |
[[Image:model equation.png|Tri-Stable Mathematical Model]] | [[Image:model equation.png|Tri-Stable Mathematical Model]] | ||
Revision as of 19:24, 24 October 2007

|
||||||||||||||||||