Princeton/lab/bioinformatics
From 2007.igem.org
(Difference between revisions)
| Line 3: | Line 3: | ||
==Biomarkers== | ==Biomarkers== | ||
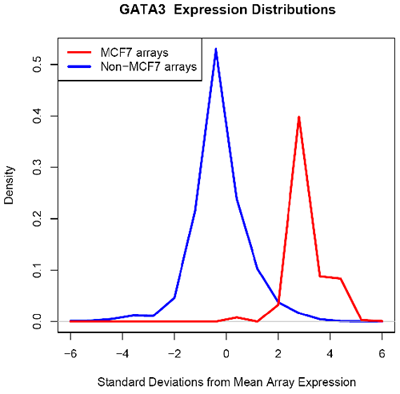
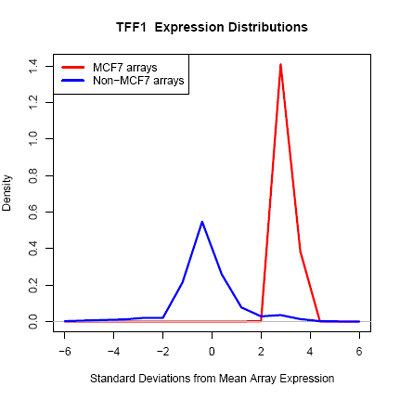
| - | Using microarray data provided by [http://www.cs.princeton.edu/~mhibbs/ Matthew Hibbs] of the [http://function.princeton.edu/ Function Group] at the [http://genomics.princeton.edu/ Lewis-Sigler Institute for Integrative Genomics], [http://www.princeton.edu/ Princeton University], it is noted that several factors, including GATA3, NPY1R | + | Using microarray data provided by [http://www.cs.princeton.edu/~mhibbs/ Matthew Hibbs] of the [http://function.princeton.edu/ Function Group] at the [http://genomics.princeton.edu/ Lewis-Sigler Institute for Integrative Genomics], [http://www.princeton.edu/ Princeton University], it is noted that several factors, including GATA3, NPY1R, and TFF1, are likely overexpressed across 14000 microarray conditions in MCF-7 cells. In particular, GATA3 in MCF-7 is likely overexpressed in 67 of 70 microarrays. |
The overexpressed factors may be used in conjunction in the AND circuit to provide greater specificity or the AND OR circuit to provide greater sensitivity. | The overexpressed factors may be used in conjunction in the AND circuit to provide greater specificity or the AND OR circuit to provide greater sensitivity. | ||
| Line 12: | Line 12: | ||
[[Image:Princeton_microarray_mcf3_npy1r.png|center|frame|Figure 3. image: Expression distribution of NPY1R in MCF-7 cells; microarray data from Matthew Hibbs.]] | [[Image:Princeton_microarray_mcf3_npy1r.png|center|frame|Figure 3. image: Expression distribution of NPY1R in MCF-7 cells; microarray data from Matthew Hibbs.]] | ||
| - | |||
| - | |||
Revision as of 22:48, 25 October 2007
Bioinformatics
Biomarkers
Using microarray data provided by [http://www.cs.princeton.edu/~mhibbs/ Matthew Hibbs] of the [http://function.princeton.edu/ Function Group] at the [http://genomics.princeton.edu/ Lewis-Sigler Institute for Integrative Genomics], [http://www.princeton.edu/ Princeton University], it is noted that several factors, including GATA3, NPY1R, and TFF1, are likely overexpressed across 14000 microarray conditions in MCF-7 cells. In particular, GATA3 in MCF-7 is likely overexpressed in 67 of 70 microarrays.
The overexpressed factors may be used in conjunction in the AND circuit to provide greater specificity or the AND OR circuit to provide greater sensitivity.