Caltech/Project/Riboregulator Results
From 2007.igem.org
| Line 12: | Line 12: | ||
==Quanta Data and Analyses== | ==Quanta Data and Analyses== | ||
Cis repression has worked on every construct tested so far, reducing expression to nearly background levels. Trans activation has been tested for cis3 and cis4 (with their trans combinations 1 and 2), as well as trans3 with cis8, making the total count: | Cis repression has worked on every construct tested so far, reducing expression to nearly background levels. Trans activation has been tested for cis3 and cis4 (with their trans combinations 1 and 2), as well as trans3 with cis8, making the total count: | ||
| - | * | + | * cis3/trans1 |
| - | * | + | * cis3/trans2 |
| - | * | + | * cis4/trans1 |
| - | * | + | * cis4/trans2 |
| - | * | + | * cis8/trans3 |
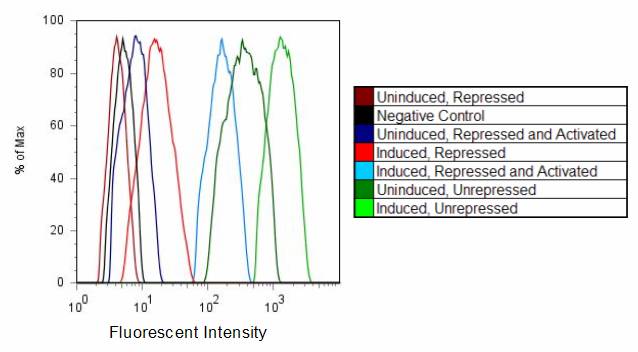
Riboregulation was tested using YFP under the control of the tet-inducible system described [[Caltech/Project/Constructs|here]]. The appropriate cis repressors were cloned in, replacing the spacer region. Those cells were cotransformed with plasmids containing the trans-activating RNA. Cells were prepared according to the [[IGEM:Caltech/2007/Protocols/Flow_Cytometry |flow cytometry]] protocol, and YFP fluorescence was measured by flow cytometry, as shown below. Arabinose is added to the trans samples to induce trans production, and aTc added to increase production of the cis transcript. | Riboregulation was tested using YFP under the control of the tet-inducible system described [[Caltech/Project/Constructs|here]]. The appropriate cis repressors were cloned in, replacing the spacer region. Those cells were cotransformed with plasmids containing the trans-activating RNA. Cells were prepared according to the [[IGEM:Caltech/2007/Protocols/Flow_Cytometry |flow cytometry]] protocol, and YFP fluorescence was measured by flow cytometry, as shown below. Arabinose is added to the trans samples to induce trans production, and aTc added to increase production of the cis transcript. | ||
Latest revision as of 01:52, 27 October 2007
iGEM 2007
Home
Highlights
Project
People
Protocols
Quanta Data and AnalysesCis repression has worked on every construct tested so far, reducing expression to nearly background levels. Trans activation has been tested for cis3 and cis4 (with their trans combinations 1 and 2), as well as trans3 with cis8, making the total count:
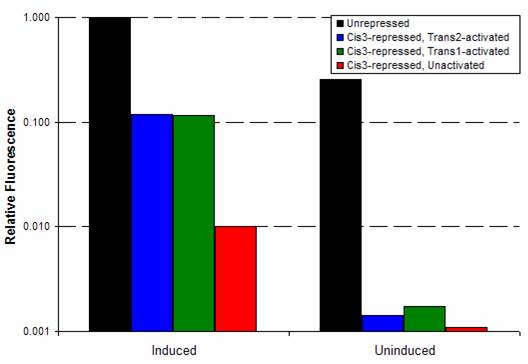
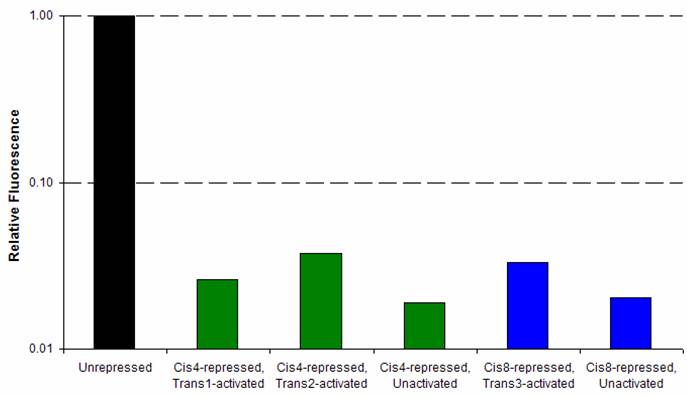
Riboregulation was tested using YFP under the control of the tet-inducible system described here. The appropriate cis repressors were cloned in, replacing the spacer region. Those cells were cotransformed with plasmids containing the trans-activating RNA. Cells were prepared according to the flow cytometry protocol, and YFP fluorescence was measured by flow cytometry, as shown below. Arabinose is added to the trans samples to induce trans production, and aTc added to increase production of the cis transcript. The fluorescence levels of D1210 (the negative control), the unrepressed/uninduced YFP construct, and YFP construct + aTc serve as a controls. Titering experience has shown that unrepressed/uninduced levels are sufficient for lysis. Therefore, the YFP fluorescence levels provide a estimate of target trans activation levels. Cis3Geometric mean fluorescences were calculated using FlowJo software and are shown below. The background autofluorescence was subtracted from each value, except for the cis3-repressed, uninduced sample (in which the fluorescence was below background). Each construct was compared to the induced, unrepressed YFP fluorescence. Trans1 and trans2 both showed ~12-fold activation, from 1% to 12% of unrepressed expression. Trans-activated, cis3-repressed constructs were roughly half as the uninduced, unrepressed YFP (and the uninduced, unrepressed Q and N expression levels are sufficient for lysis). Cis4 and Cis8Again, cis repression was relatively tight for both cis4 and cis8, roughly 2% of unrepressed expression. Trans activation, however, is not as high. At best, for cis4 and trans2, the activation was only 2-fold, from 2% to 4% of unrepressed expression. SummaryThe most promising results are for cis3 and its trans combinations, or cis3/trans1 and cis3/trans2. Both of these combinations show significant trans activation. We are currently transforming the trans1 and trans2 plasmids into the cis3-containing Q construct. Hopefully, trans activation levels are enough to give plaques for the cis3/trans1 and cis3/trans2 combinations, thus showing that Q levels are high enough for successful phage infection. |