Princeton/lab/bioinformatics
From 2007.igem.org
| Line 2: | Line 2: | ||
<html> | <html> | ||
<img src="http://img341.imageshack.us/img341/8195/logo5oz0.jpg" alt="Information"> | <img src="http://img341.imageshack.us/img341/8195/logo5oz0.jpg" alt="Information"> | ||
| - | </html> | + | </html> [[Princeton/overview | Overview]] || [[Princeton/People | People ]] || [[Princeton/lab | Lab work]] || [[Princeton/lab/experimentation | Experiments]] || [[Princeton/lab/bioinformatics | Bioinformatics]] || [[Princeton/lab/simulation | Simulations]] |
=Bioinformatics= | =Bioinformatics= | ||
Latest revision as of 02:50, 27 October 2007
 Overview || People || Lab work || Experiments || Bioinformatics || Simulations
Overview || People || Lab work || Experiments || Bioinformatics || Simulations
Contents |
Bioinformatics
Biomarkers
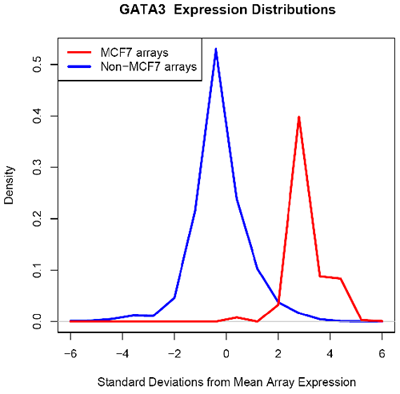
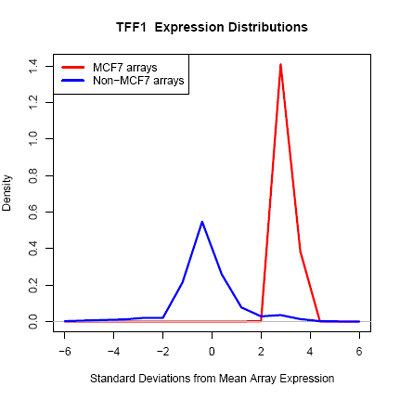
Using microarray data provided by [http://www.cs.princeton.edu/~mhibbs/ Matthew Hibbs] of the [http://function.princeton.edu/ Function Group] at the [http://genomics.princeton.edu/ Lewis-Sigler Institute for Integrative Genomics], [http://www.princeton.edu/ Princeton University], it is noted that several factors, including GATA3, NPY1R, and TFF1, are likely overexpressed across 14000 microarray conditions in MCF-7 cells. In particular, GATA3 in MCF-7 is likely overexpressed in 67 of 70 microarrays.
The overexpressed factors may be used in conjunction in the AND circuit to provide greater specificity or the AND OR circuit to provide greater sensitivity.
Design
General
[http://www.invitrogen.com/ Invitrogen] Vector NTI is used as a tool to design our constructs, primers, and restriction maps and to mark annotations. Although [http://www.ncbi.nlm.nih.gov/Sitemap/samplerecord.html GenBank format files] are available for export from [http://www.invitrogen.com/ Invitrogen] Vector NTI, the [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2007&group=Princeton Registry submissions] are in the format requested.
siRNA
[http://i.cs.hku.hk/~sirna/software/sirna.php siRNA Design Software], a meta-design tool, is utilized with mutated input sequences for imperfect binding to GATA3. The mutated input sequences are produced by introducing point mutations (at 1+j*n where j = 21 and n = 0 to the length of GATA3) that have been suggested by literature to most likely be effective.
Sequence verification
For verification of our sequencing results, an implementation of [http://www.ebi.ac.uk/Tools/clustalw/index.html ClustalW] within [http://www.invitrogen.com/ Invitrogen] Vector NTI and [http://www.ncbi.nlm.nih.gov/BLAST/ BLAST] are used for sequence comparison while [http://www.geospiza.com/finchtv/ FinchTV] is used to examine the chromatography trace data associated with sequencing.