Caltech/Project/Riboregulator
From 2007.igem.org
| Line 33: | Line 33: | ||
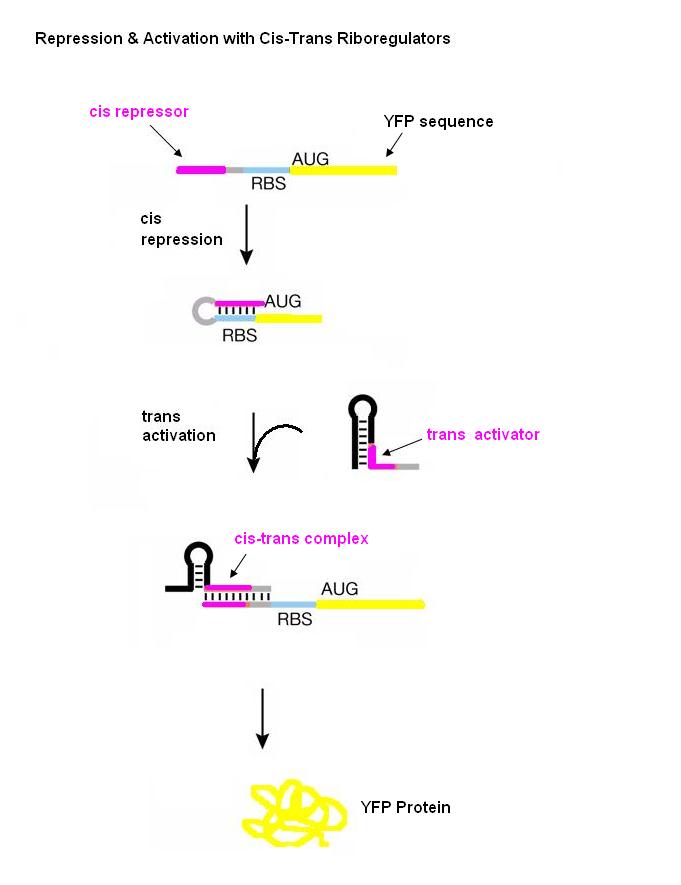
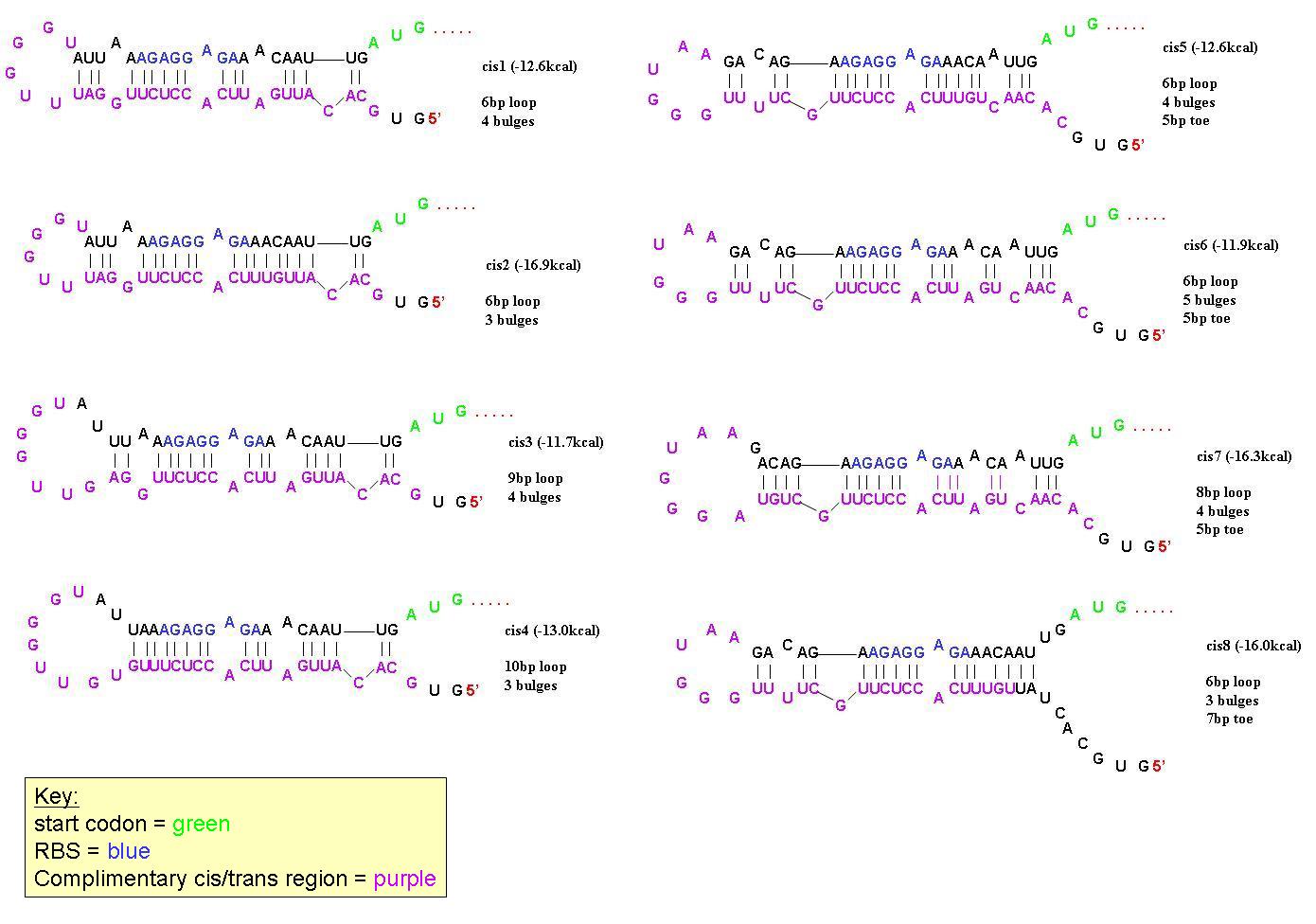
In order to find a riboregulator with the necessary dynamic range, a total of eight cis repressive elements were designed (cr1-cr8). These consisted of the same overall stem-loop structure, but had varying strengths of complementarity within the stem, as seen by their free energies. The designs of cr1-cr4 were based on previous work done by Collins et. al. and those for cr5-cr8 were based on work done by the University of California Berkeley 2006 International Genetically Engineered Machines team. | In order to find a riboregulator with the necessary dynamic range, a total of eight cis repressive elements were designed (cr1-cr8). These consisted of the same overall stem-loop structure, but had varying strengths of complementarity within the stem, as seen by their free energies. The designs of cr1-cr4 were based on previous work done by Collins et. al. and those for cr5-cr8 were based on work done by the University of California Berkeley 2006 International Genetically Engineered Machines team. | ||
| - | In order to determine the optimal level of repression, various aspects of RNA secondary structure were considered, including inner loops, single base pair bulges, and varying loop sizes. Higher free energies (i.e. less complementarity due to base pair mismatches) favor activation by taRNA because they destabilize the stem and facilitate the open RBS form upon addition of taRNA. The cis elements were inserted downstream of the Ptet promoter. The stem consisted of approximately 20-nt and the loop ranged from 6 to 10-nt. The YFP gene was inserted directly downstream of the cis sequence. Flow cytometry measurements were taken to quantify the expression of the cis riboregulated YFP gene. | + | In order to determine the optimal level of repression, various aspects of RNA secondary structure were considered, including inner loops, single base pair bulges, and varying loop sizes. Higher free energies (i.e. less complementarity due to base pair mismatches) favor activation by taRNA because they destabilize the stem and facilitate the open RBS form upon addition of taRNA. The cis elements were inserted downstream of the Ptet promoter. The stem consisted of approximately 20-nt and the loop ranged from 6 to 10-nt. The YFP gene was inserted directly downstream of the cis sequence. Flow cytometry measurements were taken to quantify the expression of the cis riboregulated YFP gene. |
| + | [[Image:Cis designs 1.JPG|center|thumb|700px|All 8 cis-repressive riboregulators have similar structure but differ in the number of base-pairs in end-loops, number of bulges, and number of base pairs on the 5' toe. They were designed this way to find the optimal dynamic range in repression/activation of protein expression.]] | ||
===Trans Activation=== | ===Trans Activation=== | ||
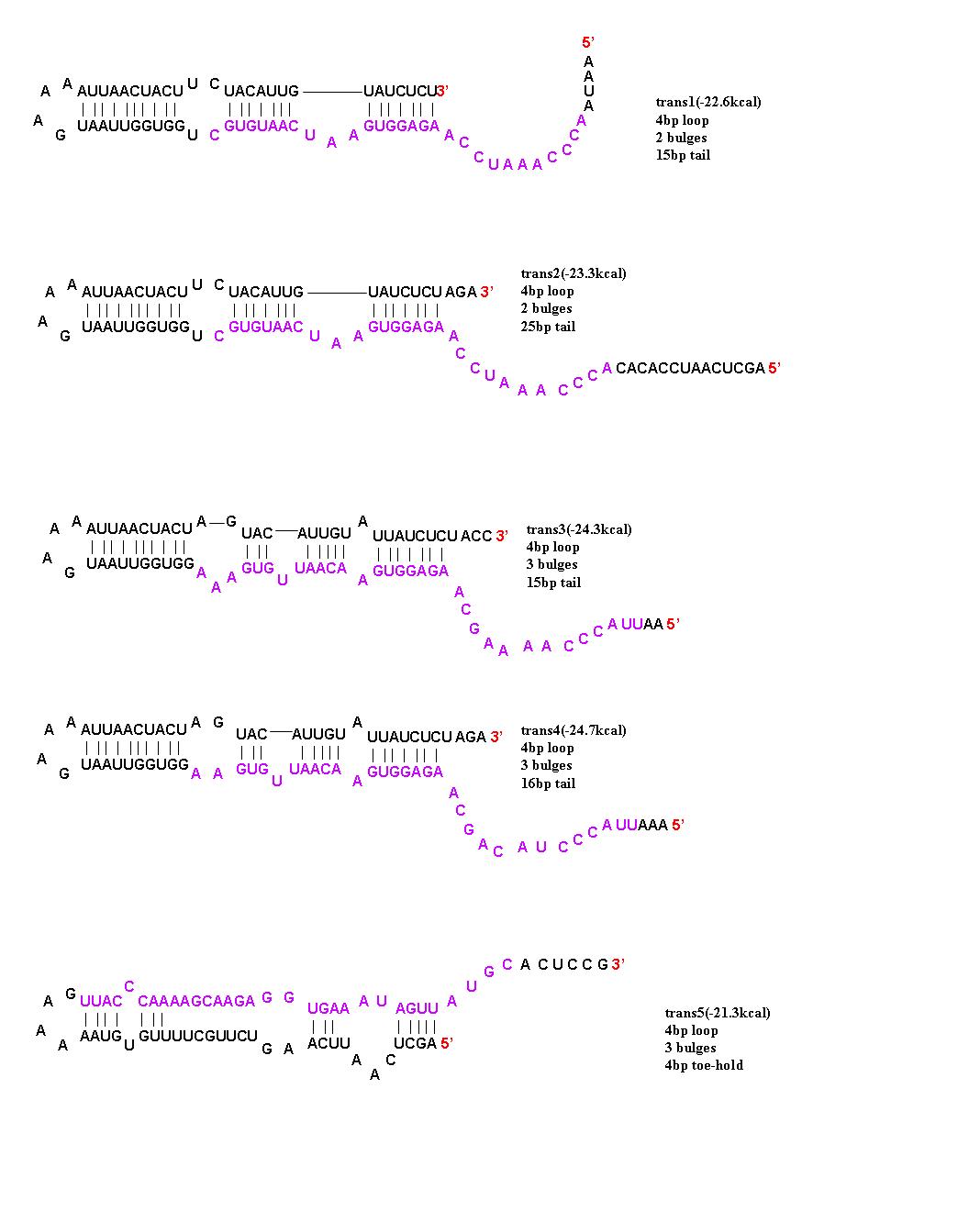
To initiate translation, five trans-activating elements (ta1-ta5) were designed. Each sequence binds to the cis-repressive elements and opens up the RBS. For ta1-ta4, part of the sequence was complementary to the hairpin loop of the cr element and extending in the 5’ direction. Element ta5 was designed to bind to the 5’ end of the cis regulator and open up the hairpin in the 3’ direction. trans1 and trans2 are complimentary to cis1-cis4; trans3 is complimentary to cis5, cis6, and cis8; trans4 is complimentary to cis7; and trans5 is complimentary to cis5-8. The activation with these ta elements is currently being [[Caltech/Project/Riboregulator_Results|determined]]. | To initiate translation, five trans-activating elements (ta1-ta5) were designed. Each sequence binds to the cis-repressive elements and opens up the RBS. For ta1-ta4, part of the sequence was complementary to the hairpin loop of the cr element and extending in the 5’ direction. Element ta5 was designed to bind to the 5’ end of the cis regulator and open up the hairpin in the 3’ direction. trans1 and trans2 are complimentary to cis1-cis4; trans3 is complimentary to cis5, cis6, and cis8; trans4 is complimentary to cis7; and trans5 is complimentary to cis5-8. The activation with these ta elements is currently being [[Caltech/Project/Riboregulator_Results|determined]]. | ||
| - | [[Image: | + | [[Image:Trans designs 1.JPG|center|thumb|700px|The 5 trans-activating riboregulators are shown with purple bases indicating regions of complimentarity to the cis-repressive riboregulators.]] |
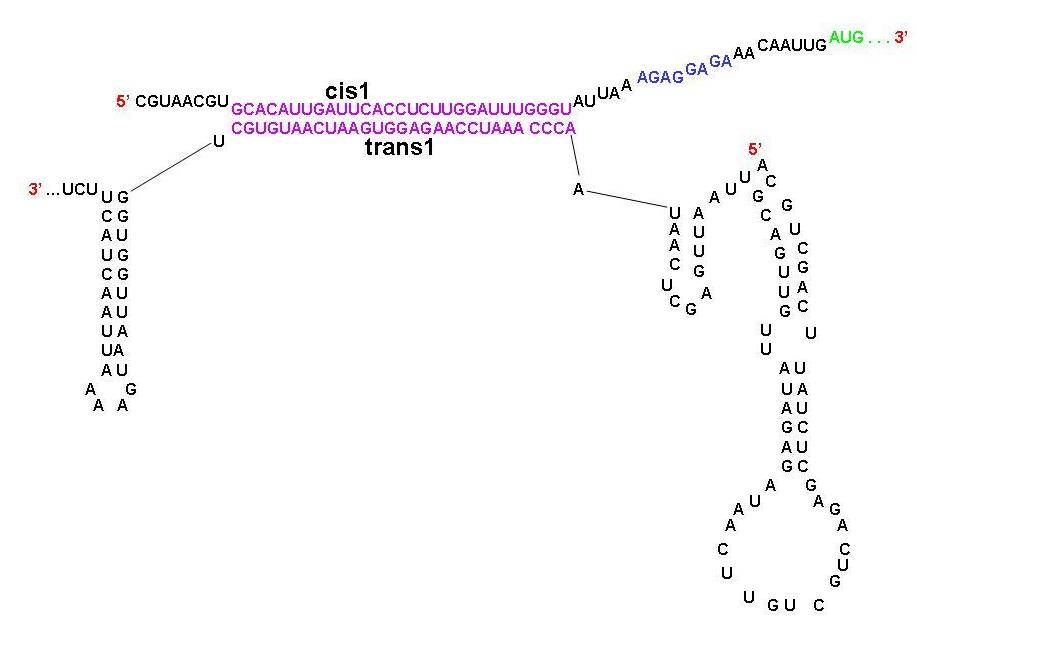
| - | + | ===Cis-trans combinations=== | |
| - | + | [[Image:Cis trans compliments.JPG|center|thumb|700px|A schematic diagram showing the parts of cis1 repressor and trans1 activator that are complimentary to each other. Note that the RBS is now open.]] | |
| - | + | ||
| - | + | ||
| - | [[Image:Cis trans compliments.JPG|center| | + | |
==Experiments with Riboregulators== | ==Experiments with Riboregulators== | ||
| Line 54: | Line 52: | ||
==Status and Future Plans== | ==Status and Future Plans== | ||
Please see our [[Caltech/Project/Riboregulator_Results|results page]] for the current status of these experiments. | Please see our [[Caltech/Project/Riboregulator_Results|results page]] for the current status of these experiments. | ||
| + | |||
|} | |} | ||
</div> | </div> | ||
Latest revision as of 03:46, 27 October 2007
iGEM 2007
Home
Highlights
Project
People
Protocols
|