Chiba/Engeneering Flagella
From 2007.igem.org
(→Results&Discussion) |
(→Results&Discussion) |
||
| Line 64: | Line 64: | ||
====Results&Discussion==== | ====Results&Discussion==== | ||
| - | '''1.Stickiness check using FliC strain''' | + | '''1.''Stickiness'' check using FliC strain''' |
[[Image:Chiba Beads-Adsorption result3.png|frame|left|fig2. Strain JW1908(''⊿fliC'',).]]<br clear="all"> | [[Image:Chiba Beads-Adsorption result3.png|frame|left|fig2. Strain JW1908(''⊿fliC'',).]]<br clear="all"> | ||
*Cell without His-FliC bound better to the Beads? No way! | *Cell without His-FliC bound better to the Beads? No way! | ||
| - | *We thought the problem might be the super-fast revolution of flagella itself. We decided to try MotB strain.<br> | + | *We thought the problem might be the super-fast revolution of flagella itself. We decided to try MotB strain.<br><br><br> |
[[Image:Chiba_Beads-Adsorption_result2.png|frame|left|fig3. Strain GI826(''⊿fliC'',''⊿motB'').]]<br clear="all"> | [[Image:Chiba_Beads-Adsorption_result2.png|frame|left|fig3. Strain GI826(''⊿fliC'',''⊿motB'').]]<br clear="all"> | ||
Revision as of 04:46, 27 October 2007
|
Introduction | Project Design ( 1.Affinity Tag | 2.Communication Module | 3.Size Control ) | Making Marimos | Our Goal |
Stickey Tags
Our Aim
To make stickey hands on E.coli, we focused on their flagella that are located outside the cells. We used the following mechanisms:
- Display sticky peptides in flagellar filament.
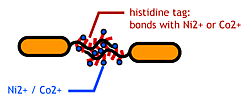
- His-tag. The imidazole group in histidines make a complex with metal ions.
We combined these two and made a His-tagged flagella in the hope to stick them together via metal ions.
[http://www.npn.jst.go.jp/index.html About flagella]
E.Coli have 5-10 flagella. The flagella is used for swimming and for chemotaxis; the bacteria run when they find attractant, tumble when there is a repellent.
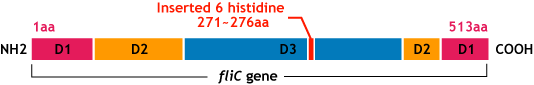
E.coli flagella consist of three parts: a basal body, a hook, and a filament. The filament of E.Coli is a rigid, helical, and cylindrical structure which is 10-15μm long and 23nm thick in diameter. It is built from ~20000 subunits of a ~55kDa single protein, FliC. FliC has three domains, D1,D2,D3; although D1 and D2 are needed for the formation of the functional flagellar filament, D3 domain which sticks outside of the fillament are not essential[3].
"Variable" FliC D3 domain
It is reported that the proteins up to 49.4kDa could be displayed on the cell surface of E.Coli using flagellin fusion protein.[4]
About Histidine Tag
See [http://en.wikipedia.org/wiki/His-tag wikipedia article].
Experiments
Making FliC-his gene
- We inserted the short peptide with six histidine (“His-Tag”) into the fliC D3 domain.
Checking the "Stickiness": Beads Adsorption
Purpose
Confirm that the his-tags are displaied on the flagella and are capable of binding to Co2+- or Ni2+- surface.
Samples
- ⊿fliC strain(JW1908 in KEIO collections [5]) transformed with
- pUC19-fliC-his
- no plasmid
- ⊿fliC,⊿motB strain(GI826)transformed with
- pUC19-fliC-his
- no plasmid
Testing Procedure
- pUC19-FliC-His was transformed to JW1908(fliC) and GI826(fliC motB).
- Grown to stationary phase
- Culture suspended with Dynabeads (Metal-IDA), allowing to the affinity adsorption
- Beads washed with a phosphate buffer (x4)
- E" coli" detached from beads by adding imidazole then spreaded on agar plates.
- The number of the colonies on resultant plates.
Results&Discussion
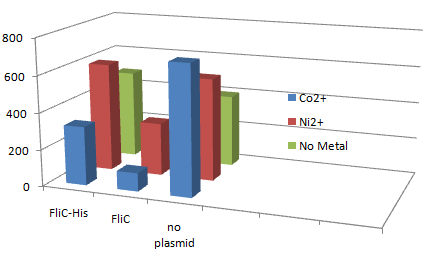
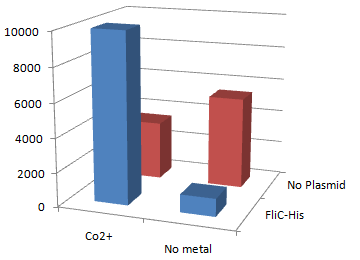
1.Stickiness check using FliC strain
- Cell without His-FliC bound better to the Beads? No way!
- We thought the problem might be the super-fast revolution of flagella itself. We decided to try MotB strain.
- In the presence of Co2+Histidine tag,beads adsorb bacteria.
Co2+Histidine Tagの存在よって大腸菌がビーズに吸着している.
- In the presence of FliC-His, cobalt ion adsorb bacteria stronger than nickel ion.
- Ni2+よりもCo2+のほうがfliC-his存在下でより吸着している.
- Ni2+よりもCo2+のほうがfliC-hisの有無で吸着の差が大きい
- The number of colony dramatically decreased with out Co2+ or FliC-His plasmid.
2.Strainの比較
- 鞭毛の回転がなければある程度の吸着力を保つ。しかしワイルドタイプのFliCの発現も伴うため、Histidineによる吸着度合いは低下してしまう。
3.金属イオンの比較 一般的に知られているように、コバルトのほうがHistidineとより結合をつくるのではないか。
References
3. Kuwajima, G. et al.: J. Bacteriol., 170, 3305-3309 (1988)
4. Ezaki, S. et. al.: J. Ferment. Bioeng., 86, 500-503 (1998)
5. Baba, T. et. al.: Mol. Systems. Biol., 21, 1-10 (2006)