Chiba/Engeneering Flagella
From 2007.igem.org
(→Stickey Tags) |
(→Our Aim) |
||
| Line 11: | Line 11: | ||
===Our Aim=== | ===Our Aim=== | ||
[[Image:Chiba_stickbacteria.png|frame|'''Fig. 9''' Bacteria Linker]] | [[Image:Chiba_stickbacteria.png|frame|'''Fig. 9''' Bacteria Linker]] | ||
| - | To make | + | To make affinity tags on ''E.coli'', we focused on their flagella that are located outside the cells. We used the following mechanisms: |
*Display sticky peptides in flagellar filament. | *Display sticky peptides in flagellar filament. | ||
*His-tag. The imidazole group in histidines make a complex with metal ions. | *His-tag. The imidazole group in histidines make a complex with metal ions. | ||
Revision as of 05:09, 27 October 2007
|
Introduction | Project Design ( 1.Affinity Tag | 2.Communication Module | 3.Size Control ) | Making Marimos | Our Goal |
Affinity Tags
Our Aim
To make affinity tags on E.coli, we focused on their flagella that are located outside the cells. We used the following mechanisms:
- Display sticky peptides in flagellar filament.
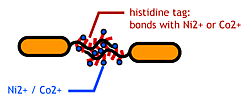
- His-tag. The imidazole group in histidines make a complex with metal ions.
We combined these two and made a His-tagged flagella in the hope to stick them together via metal ions.
[http://www.npn.jst.go.jp/index.html About flagella]
E.Coli have 5-10 flagella. The flagella is used for swimming and for chemotaxis; the bacteria run when they find attractant, tumble when there is a repellent.
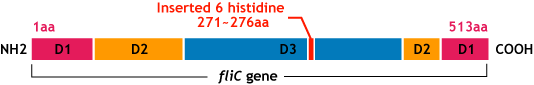
E.coli flagella consist of three parts: a basal body, a hook, and a filament. The filament of E.Coli is a rigid, helical, and cylindrical structure which is 10-15μm long and 23nm thick in diameter. It is built from ~20000 subunits of a ~55kDa single protein, FliC. FliC has three domains, D1,D2,D3; although D1 and D2 are needed for the formation of the functional flagellar filament, D3 domain which sticks outside of the fillament are not essential[3].
"Variable" FliC D3 domain
It is reported that the proteins up to 49.4kDa could be displayed on the cell surface of E.Coli using flagellin fusion protein.[4]
About Histidine Tag
See [http://en.wikipedia.org/wiki/His-tag wikipedia article].
Experiments
Making |frame|FliC-his gene
- We inserted the short peptide with six histidine (“His-Tag”) into the fliC D3 domain.
Checking the "Stickiness": Beads Adsorption
Purpose
Confirm that the his-tags are displaied on the flagella and are capable of binding to Co2+- or Ni2+- surface.
Samples
- ⊿fliC strain(JW1908 in KEIO collections [5]) transformed with
- pUC19-fliC-his
- no plasmid
- ⊿fliC,⊿motB strain(GI826)transformed with
- pUC19-fliC-his
- no plasmid
Testing Procedure
- pUC19-FliC-His was transformed to JW1908(fliC), JW0747(MotB), and GI826(fliC motB).
- Grown to stationary phase
- Culture suspended with Dynabeads (Metal-IDA), allowing to the affinity adsorption
- Beads washed with a phosphate buffer (x4)
- E" coli" detached from beads by adding imidazole then spreaded on agar plates.
- The number of the colonies on resultant plates.
Results&Discussion
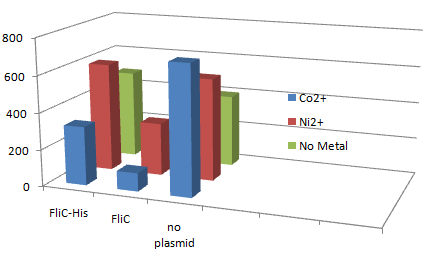
1.Stickiness check using FliC strain
- Cell without His-FliC bound better to the Beads? No way!
- We thought the problem might be the super-fast revolution of flagella itself. We decided to try MotB strain.
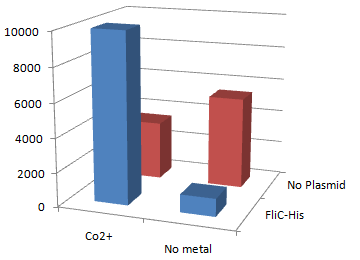
2.Stickiness check using MotB strain
- Mot B deletion provides cell with the flagella completely assembled but not rotating.
*This time everything worked out! Only in the presence of Co2+Bacteria with His-FliC sticked to the Co-IDA beads very well.
- In this strain, FliC-His is assembled with wildtype FliC coded in genomic DNA. Nevertheless, the binding efficiency was at the same level (not shown). it seems that His-Tag displayed on the flagella is enough to do its work.
- On the other hand, the deletion of MotB turned out to be vital for sticking the tagged flagella together.
- In the presence of FliC-His, cobalt ion adsorb bacteria stronger than nickel ion, this was more or less the expected result.
References
3. Kuwajima, G. et al.: J. Bacteriol., 170, 3305-3309 (1988)
4. Ezaki, S. et. al.: J. Ferment. Bioeng., 86, 500-503 (1998)
5. Baba, T. et. al.: Mol. Systems. Biol., 21, 1-10 (2006)