Vaibhavi Umesh Notebook
From 2007.igem.org
My Construction Files
My Sequencing Files
~~!~~
notes
to do
Vaibhaviu 19:02, 1 June 2007 (EDT)
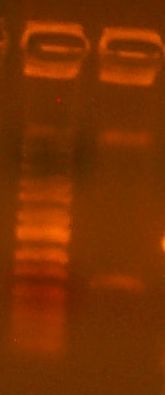
Today we cloned an RFP basic part into pBca9145. So, we did the construction file for pBca9145-Bca1145. We did a PCR of the RFP gene according to the construction file, and the gel is here:
It's a little dilute, but it should work nonetheless. We also digested the vector according to the construction file:
We gel purified the larger of the two bands. It looks like the digest went pretty well, as we see no single-cut plasmid DNA. We took it all the way through to transformation into DH10B cells today.
Vaibhaviu 18:45, 5 June 2007 (EDT)
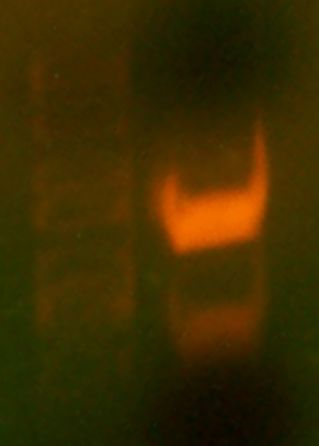
Today we first observed the transformed plates that grew over the weekend. Four large, white, colonies surrounded by several satellite colonies were observed. No red colonies were observed which meant no parent vector had bled through onto the plate. FOr each colony, we made overnight cultures and placed them in the 'Incushaker'. Chris had already simulated this and two samples of the cultures were already ready for miniprep. We simulated a mini prep experiment and isolated the plasmids from the two culture samples. We then performed the analytical test digest and used BglII and XhoI restriction enzymes. These enzymes were chosen because they gave a distinct digestion pattern when compared with the parent vector.
Then, we simulated a colony PCR on one of the colonies on the plate, chosen randomly. We ran the results of both the digest and the PCR on a gel. The gel contained 5 wells. The first one contained a marker (the ladder), the second one The results of the gel were not very clear. THe parent vector and the PCR lanes both did not show up/ did not display the expected results. THe pictures taken were unclear, but when observed on the UV plate, the bands were brighter, but no additional bands showed up.
We also made the samples to be sequenced. We placed the mini prep plasmids in a 1.5 mL epp tube. We filled out the spreadsheet to send to Quintara and placed the samples in the metal box outside to get picked up. The specific oligos to be used were also mentioned in the spreadsheet.
Vaibhaviu 18:45, 5 June 2007 (EDT)
Today, we got the sequencing results from Quintara
6/12
Today I got my sequencing results back. SodC #1: SodC #2 was fully correct. However, at base pair # ______, there was a mis-read of 3 C’s when only two distinct peaks could be seen on the graph. For methAP gene results, methAP#1 yielded a better result than methAP#2. However, although all the base pairs aligned for methAP#1, there was one situation at base pair # _____, where the sequencing result had inserted an extra G not present in the original gene. When the graph was observed, the resulting peak was quite ambiguous and it was difficult to distinguish whether the peak was actually two separate peaks, or was three. As a result, just to double check, the plasmid was sent in for reverse sequenicing using the reverse primer G00101 (the reverse primer of ca998). The results for this sequencing file will help establish that the mis-match and presence of an extra G was due to the ambiguity of the graph rather than the presence of an actual mutation.
VU005.MetAP#1 is the sample I sent in along with the primer G00101