|
Tri-stable Toggle Switch
The purpose of the Tri-stable Toggle Switch is to produce three distinct and stable outputs in response to three distinct inputs. These three inputs are three separate chemicals which will each induce one state of the switch. 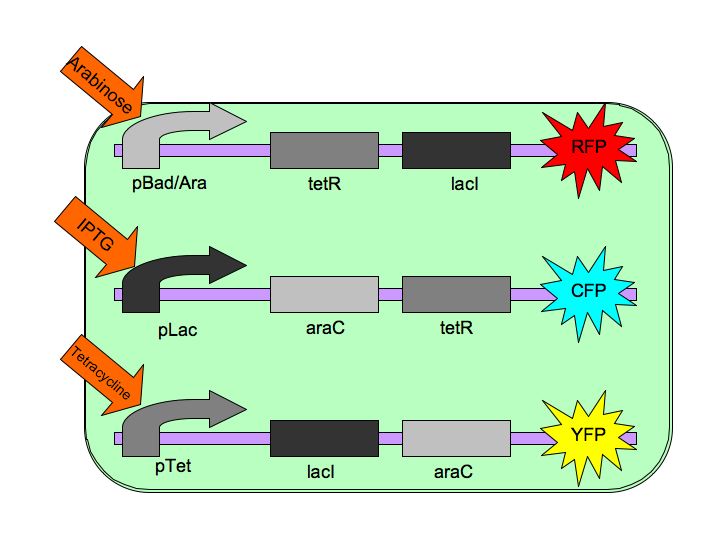 The Tri-stable Toggle Switch Architecture pBAD->LacI->TetR,
pLacI->AraC->TetR and
pTet->AraC->LacI,
where [http://en.wikipedia.org/wiki/L-arabinose_operon AraC] represses pAraC/BAD, [http://en.wikipedia.org/wiki/Lac_repressor LacI] represses pLac and [http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation TetR] represses pTet.
Each of the three repressors are inactivated by one of three chemicals, the three inducer chemicals mentioned earlier. These three([http://en.wikipedia.org/wiki/Arabinose arabinose], [http://en.wikipedia.org/wiki/IPTG IPTG] (Isopropyl β-D-1-thiogalactopyranoside) and [http://en.wikipedia.org/wiki/Tetracycline Tetracycline], respectively), cause conformational changes in their respective repressor proteins which keeps them from binding to DNA in an inhibitory manner which leads to gene expression. For example, in the presence of arabinose, AraC cannot repress pAraC/BAD so LacI and TetR are produced which in turn repress pTet and pLac and the pAraC/BAD construct is turned on.
AraC/BAD
The gene AraC, one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html]
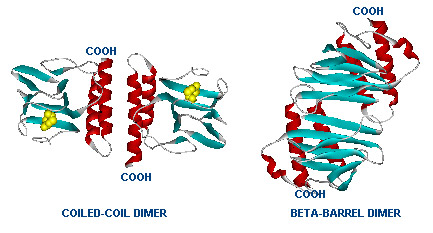 Dimer structure with arabinose on the left (yellow) 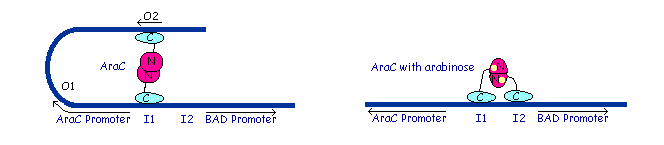 The left image shows the araC dimer repressing transcription, while the right conformation enables transcription
LacI
In nature, LacI represses pLac which promotes the LacYZA genes that metabolize lactose. Thus LacI represses pLac except in the presence of lactose (or lactose mimics, eg IPTG). 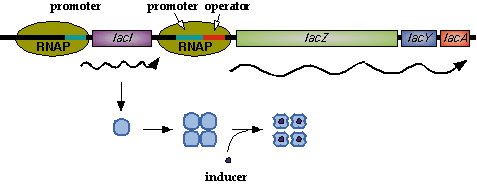 Image[http://www.mun.ca/biochem/courses/3107/Topics/Lac_genetics.html]. LacI forms a tetramer and represses pLac. However, an inducer, such as IPTG, causes a conformation change that removes LacI from the operator site.
TetR
TetR represses the constitutive promoter pTet. In the presence of tetracycline, an antibiotic, a conformational change in TetR inhibits the protein from binding to the operator region. In nature, pTet promotes TetR and TetA. The latter of which acts to pump tetracycline out of the cell, thus the pump is only activated in the presence of Tetracycline.[http://en.wikipedia.org/wiki/Tetracycline_controlled_transcriptional_activation]
The TetR, as it turns out is a very tight repressor and a range of 0 to 1 ug/ml has been shown to cause a 5 order of magnitude change in luciferase production.[http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=1319065&query_hl=1&itool=pubmed_docsum]
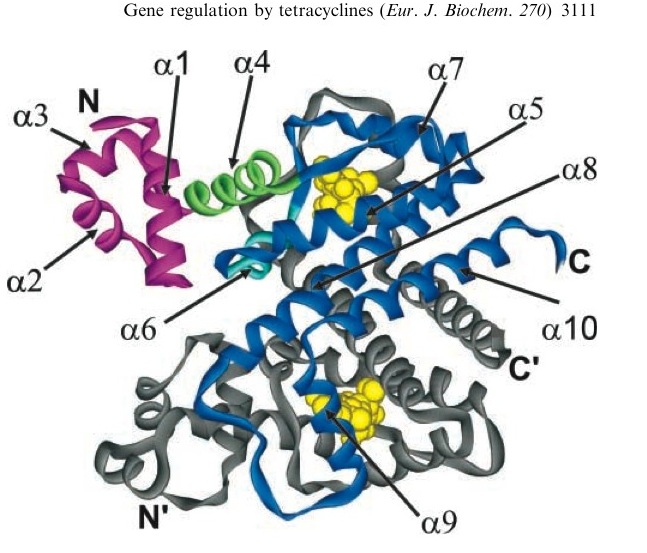 A tetracycline molecule binds to each of the two TetR monomers to form a dimer
Modeling
The first draft of the code started last year:
[http://parts2.mit.edu/wiki/index.php/Table_of_preliminary_model_constants Initial Table of Constants]
[http://parts2.mit.edu/wiki/index.php/Derivation_of_the_Model_Equations Derivation of Model Equations]
Media:tristable2006.txt This code proved to complicated to work with and a more simplified version was developed.
A simpler model based on the bistable paper was developed that takes the combined, relative transcription/translation rates into account Media:tristable2007.txt.
Parameters in the Model
Repressor Production Rate
Repressor production rate is determined by two factors:
- Promoter Strength (Transcription)
- Ribosome Binding Strength (Translation)
In the model, the total repressor production rate = alpha
The promoter strength cannot be easily changed because this would require mutations in the promoter or a different promoter/repressor combo. However, RBSs have been well characterized and the alpha parameter can be modulated by inserting different strength RBSs as determined by the model and testing.
Repressor Strength
The strength of the repressors is determined by:
- The repressor concentration, [Repressor].
- The cooperativity of repression, Beta.
In the Model, the repressor strength = [Repressor]^Beta
The Beta value is characteristic of each repressor and represents a constant that cannot be changed.
Degradation
The degradation in the model is denoted by the term "-repressor," where the degradation constant is unity because the rates are relative, rather than absolute. The next iteration of the model will include degradation constants. Currently in the registry all of the repressors have strong degradation tags. Jason Kelly made a comment on one repressor and said he was suspicious of how strong of a repression would be able to be obtained from the repessors. We need strong repression for our system, so we may need to remove the tag our system to function.
The Model
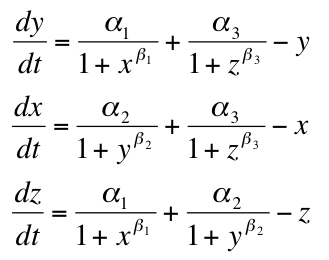
Beta = Repression Strength
Alpha = Repressor Production Rate
X = Repressor1
Y = Repressor2
Z = Repressor3
Stability

Blue points are stable combinations of repressor production rates, while red circles are unstable combinations of repressor production rates. This graph is for Beta = 2. The tristable region gets larger (i.e. more disparate alpha values will be able to constitute a tristable system) as beta gets larger. The tristable region disappears when Beta equals one or less.
An easy visual aid to seeing how increased beta values (cooperativity of repression) leads to a more stable system consider the following graph of rate of production of repressor1 vs. the repressor that inhibits the production of repressor1. dx/dt vs. [y]
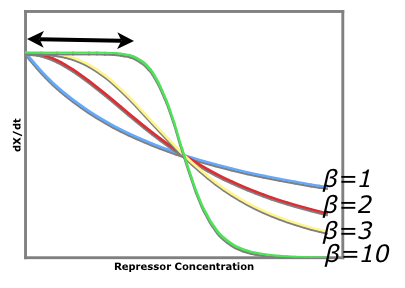
The black arrow shows that there is a larger region for larger beta values where stray repessors will not significantly affect the rate of repressor production rate whereas in a system of beta = 1, stray repressors will significantly change the system, making it less robust.
Other Predicted Obstacles
- Other iGEM teams (info via Patrick King) have found that the araC gene contains a promoter region. Grown in TOP10 cells the construct araC>RBS>GFP>Terminator was observed to give a strong florescent output. In our system, this would give a florescent output for one FP even if that construct was not on.
- AraC exhibits all or none gene expression in the presents of arabinose because the natural transporter, when induced, causes that cell to transport arabinose, depleting the culture. This may be overcome by adding a saturating level of arabinose. In the arabinose construct already made, we were not able to observe FP output even though there was a high level of arabinose. We used XL1Blue cells.
- The degradation tags on the repressors may not give a long enough protein life time to yield strong repression. Thus, the tags may need to be removed to have a working device.
|

