Austin Day Notebook
From 2007.igem.org
My Construction Files
My Sequencing Files
AustinDay 16:48, 22 June 2007 (EDT)
- So we had the great flood today. Looks like a tornado went through the offices.
- Digested, ligated, and transformed the I716067 part (added the promoter onto the 2X mutant hemoglobins)
AustinDay 20:24, 21 June 2007 (EDT)
- Designed the oligos for the Di-alpha mutant fusion protein.
- Scraped and grew up the I716067 (-rbs-2X mutant alpha-rbs-2X mutant beta)
- Grew up 3 clones from the 20X dilution of 9145-1154 #1.
- Still waiting for the hemoglobin and antibodies... then the real fun can start.
AustinDay 12:45, 20 June 2007 (EDT)
- The sequencing came back and the first 1154 part was a dud, the second was a failed read. This is a bit annoying. I'm going to make a 1000X dilution of the #1 clone and retransform it. The read came back correct for that one, although the gel wouldn't work... But perhaps there's still a good one it there someplace.
- I also made mini preps of all of the chassis plasmids. (9104, 9106, 9023, and flu/inv)
- I also digested and transformed the I716067 part (The joined rbs lib-beta-rbs lib-alpha 2X mutant part)
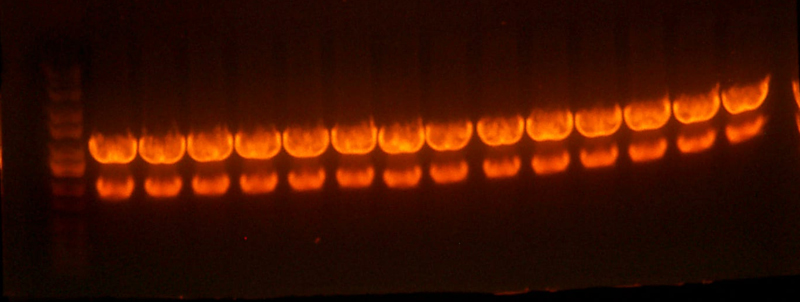
- Did a protein gel of (ladder) (9145-1089 neg control) (I716061 #1) (I716061 #2) (I716061 #3) (I716061 #4) (I716061 #10) (I716061 #11) (I716061 #12) (I716061 #14) (9145-1089)
- It appears that there is a band at about 38KDa that is much darker on the I716061 clones and very light, but I believe detectable, on the 9145-1089 negatives. That paper that did a 15% SES gel got bands for hemoglobin at ~14 and 13 KDa. There are bands that are present on all of the lanes, however, there are two which are significantly darker than any of the others, and both of those are the I716061 clones. These are in the correct position (13-14 KDa). Maybe they are different levels of hemoglobin? Chris suggests that we run a higher percentage acrylamide gel next time to be able to resolve such small bands.
AustinDay 12:20, 19 June 2007 (EDT)
- Grew up some of those I716065 and I716066 libraries.
- Grew up 9106, 9104, 9203, and flu/inv so I can make more of the mini preps.
- Ordered the hemoglobin and hemoglobin HRP conjugated primary antibody:
http://www.sigmaaldrich.com/catalog/search/ProductDetail/SIGMA/H7379
http://www.bethyl.com/abdetails.asp?antibodyid=967
- Grew up 14 clones from the I716061 plate for the SDS-PAGE gel we're hopefully going to do tomorrow.
- Also sequenced 2 more clones of Chris' 1103-1154 part.
AustinDay 14:30, 18 June 2007 (EDT)
- I digested, ligated, and transformed the 2X mutants (I716065 and I716066)
- I grew up some more of the 1154 clones chris made the other day. The sequences weren't that nice, and one of them wouldn't come out in a digest gel.
Goals for tomorrow:
- Grow up 2X mutant transformants in the morning.
- Use the 1154 and 1150 parts and put em together.
AustinDay 21:09, 14 June 2007 (EDT)
- I minied 2 of the Bca1154 and bca1150 parts that Chris made. I also minied 4 clones of the I716061 library (#2 looked the darkest, and #4 mini was diluted in 100 instead of 50ul.) I also minied 3 clones of the mutant hemoglobins. These mutations should finish up the presbyterian and providence mutation sets. I also sequenced all of them.
AustinDay 18:35, 13 June 2007 (EDT)
- So I finished up the P1 and K1E assay and tried the serum resistance assay today for the talk on saturday.
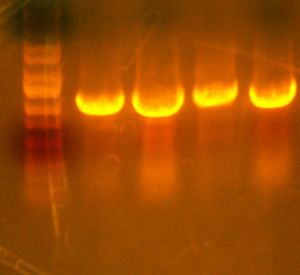
- I also grew up a few of the I716061 colonies. (They were the ones that should have the cassette with the P-trc-rbs-beta-rbs-alpha) They weren't red, but colony PCR confirmed that they have the correct size. I'm going to let the plate grow up some more and I'm going to grow up a few of them to sequence just to verify the sequence. I'm also going to look into figuring out that spec method of determining the concentration of Hb present.
- I also picked 2 of chris' plates which have the pcon-rbs-alpha and pcon-rbs-beta.
- Here's the pictures of the colony PCR thing I did for the I716061 colonies. The expected band size is 1182 bp.
AustinDay 20:06, 12 June 2007 (EDT)
- Yesterday I transformed the pTRC-rbs-beta-rbs-alpha, but I stupidly choose the wrong restriction sites. Let's just leave that behind us. Anyway, I retransformed those those today, so hopefully we'll see bloody colonies tomorrow.
- I also did the quickchange for the second set of mutations. I plated those today.
- Note to self: Check Chris' plate for hemoglobin colonies (little red ones would be ideal) And put that large taped stack of plates into the mini fridge.
- I colonies appear, pick some and grow em up. Will want to put in tandem like the format I'm working on if they work well.
AustinDay 16:16, 9 June 2007 (EDT)
- I grew up 3 SODs and 3 Met-AP genes for Vai. I also put a bunch of plates into the fridge for some other peoples.
- I minied 3 clones for the 2 hemoglobin subunit and sequenced them. I also minied the antibiotic-rbs library-hemoglobin subunit (unmutated) and digested them so that I can make that antibiotic-rbs-beta-rbs-alpha cassette.
- I also minied the 9106 plasmid of Chris' because the other one I was using for the assays for the symposium data was running low. Those strains are having a hard time transforming. I still can't get any colonies to take up the flu/inv plasmid. I think it might be running low also. Not good.
- I forgot what kind of media I grew up the p-tric transformants into yesterday. But I minied those also and made a digest before I realized that it might have been plain LB. I took a -80 stock of it and grew it up in AMP just in case.
- I also transformed more of the 9104, 9106, flu/inv, and 9023 plasmids into DH10B. Just in case I get a low supply, considering that all of the flu/inv transformations failed last time.
AustinDay 13:43, 8 June 2007 (EDT)
- The transformations of the I716056 and I716057 worked!!! I scraped em and started some cultures.
- The first quickchange reaction seemed to work. I got some colonies. I still went forward with the second quickchange reaction in case somehow these aren't the right results. I grew up 3 of each.
AustinDay 20:02, 7 June 2007 (EDT)
- Redid the digestion and ligations and transformations from yesterday. Which was remaking the I716055 and I716056 (alpha and beta subunits, unmutated, in front of the RBs library and different antibiotics)
- I also realized that I added too much oligo to my quickchange reaction yesterday, so the transformation I did from that may not have worked. I'm redoing the PCR today and running it overnight. Hopefully either the one from yesterday would work, or the one I started again today will work.
AustinDay 15:12, 6 June 2007 (EDT)
- The transformations all ... failed. Not sure why, but I did a lot of them yesterday and they all didn't really work. I'm guessing it's something that was consistently wrong, so I'm going to double check all the temperatures and reagents and try them again. I did the transformation this morning, so hopefully I can get some colonies and grow up the library tonight so that I don't really waste a day because of this.
AustinDay 15:17, 5 June 2007 (EDT)
- Designed the oligos for the hemoglobin mutants. Here are my notes on the mutants:
Primary Mutants:
Wild Type Hemoglobin
PPDA: P50 = 44, yield = 17.5%
Presbyterian mutation (Increase P50): Beta: Asn-108 -> Lys
Providence mutation (Increase yield): Beta: Lys-82 -> Asp
Di-alpha fusion (Increase yeidl): Glycine? linker.
P2996: P50 = 34, yield “average”
Presbyterian mutation (Increase P50): Beta: Asn-108 -> Lys
αL29F (Decrease P50, decrease auto-oxidation)
αV96W (Increase P50, increase auto-oxidation)
Oligo design for the following mutations:
ASN: AAC -> AAA
B-Asn108Lys
Lys AAG 14043.00 10.26 0.23
Lys AAA 46044.00 33.66 0.77
GAATTTTCGTCTGCTGGGTAAAGTGCTGGTGTGTGTCCTTGC
LYS: AAA -> GAT
B-Lys82Asp
Asp GAT 44103.00 32.24 0.63
Asp GAC 26201.00 19.15 0.37
CTGGCGCATCTTGATAATCTTGATGGTACATTCGCGACTCTGAG
Di-alpha fusion: Oligos coming soon.
LYS: TTA -> TTT
A-L29F
Phe TTT 30361.00 22.19 0.57
Phe TTC 22649.00 16.56 0.43
GAGTATGGTGCTGAAGCTTTTGAACGCATGTTTTTAAGCTTTC
VAL: GTT -> TGG
A-V96W
Trp TGG 20835.00 15.23 1.00
CGCATAAACTCCGTGTGGACCCGTGGAACTTTAAACTGCTGTCCCACTG
Secondary Mutants (Or mutations that would lead to them)
Alpha; VAL E11 -> Threonine: Increases P50 by 4x (to 25 torr). Due to stabilization of water molecule in distal heme pocket of decoy T-state. Increases auto-oxidation rate.
Beta; VAL E11 -> Threonine: Increases P50 by 2x (to 12 torr).
Alpha; V26T: 4x increase in P50. Lost cooperativity. (n = 1.1)
Beta; B67T: (n = 2.2) Slightly increased P50.
Combination of surface modifications to increase P50 by 3x:
o Val1 -> Met:
o His2 -> deleted
o Thr4 -> Ile
o Pro5 -> Ala
o Ala 76 -> Lys
βN108D, (In place of presbyterian) increases the P50 more than the presbyterian.
- In other news: All but I716053 #1 came out white. That colony was red, so I didn't mini it. (parent vector)
- I'm going to go on with the construction now with the RBs libraries from yesterday.
- I used the #1 clone from each of the parts for the next digest/ligation/transformation round.
- I transformed the colonies with the RBS library. I forgot to select for the secondary antibiotic, but Chris said I should just scrape the plate tomorrow and grow it all up in the antibiotic marker. Hopefully that'll do the trick.
AustinDay 13:25, 4 June 2007 (EDT)
- Got a nice number of colonies on the synthetic gene biobrick transformations from yesterday. I grew up 2 of each.
- I minied the RBS library parts (I716051 and I716052).
AustinDay 16:50, 3 June 2007 (EDT)
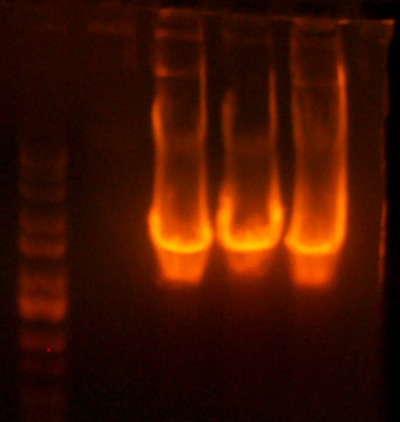
- Made an EcoRI / XhoI digest of the synthetic genes and a digest of EcoRI and XhoI of the 9145 plasmid.
- Here's the gel. Very pretty.
- Took the small fragment from the synthetic genes and the larger fragment from the 9145 plasmid and ligated them together.
- I think I'm going to name these new parts: I716053 (9186 + 9145), I716054 (9187 + 9145), I716055 (9188 + 9145). These names came to me in a dream. It was said they shall bring luck and prosperity to all who use them.
- I grew up 2 colonies of the homogenized RBS pools. (The Kan and the Cmr pools)
Austinday 15:43, 2 June 2007 (PDT)
- Okay, so since the 1121 part was bad, chris found a PCR product of it. I'm going to give that a try and see if it works.
- I did the ligation and transformation for those. I named the parts Bad0001 (kan) and Bad0002 (Cmr). Plated and cooking for tomorrow.
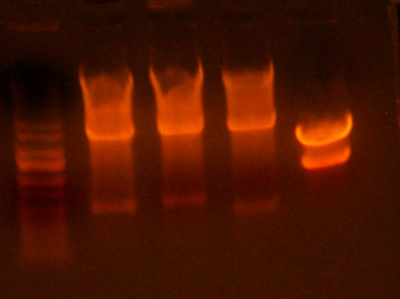
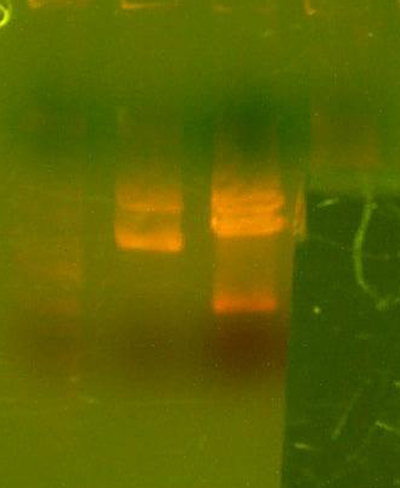
- Made digest of BglII and XhoI of the 3X concentrated 9145 plasmid miniprep. The total digest was equal to that of 14 regular digests. The gel is below:
- Digested the DNA 2.0 plasmids with EcoR1 and XhoI. But...the gel looks funny. I may not have allowed enough time for the lyophilized DNA to redissolve... That's my best guess at least.
- I grew up some colonies of the DNA 2.0 parts so I'll give it another try tomorrow. (Chris told me that there is probably too much DNA and that if I ran it longer, it might have cleared up. You can kind of see that there might be a second band...sort of.)
Austinday 22:59, 30 May 2007 (PDT)
- Okay, I repeated the digestion from yesterday, but got the same results. Here's the gel.
- This time, I let it digest for well over an hour and mixed it half way through. I also ran the gel at a lower voltage than usual.
- The bands for the 1122 lane was noticeably cleaner, but the 1121 lane was uncut.
- I cut out the smaller band from the 1122 lane(because it's probably a higher yield than the previous gel), as well as the 1128 band. I figured that because the previous gel didn't cut completely, and because the 1128 pool smaller bands would have already ran off the gel, this second gel would have a more pure band than the first. I also cut out the region of the 1121 lane where the smaller band should be, although it looks as if nothing is there... I guess we'll have to remake that 1121 part.
Austinday 16:53, 29 May 2007 (PDT)
- Sequencing of the pBca1101-Bca1128 RBS library resulted in 2 duplicate RBS's. #24 was identical to #3, and #5 was identical to #2 so I threw out #3 and #5.
- I pooled together the bca1128 rbs's and the 1106 A and B rbs's as well as one other 1128 A rbs Chris had.
- I digested those with EcoR1 and BglII to create the vectors to insert the antibiotic markers. After digestion, the pool looked good so I cut it out right away. The 1121 and 1122 (CmR and Kan) digestions didn't look correct.
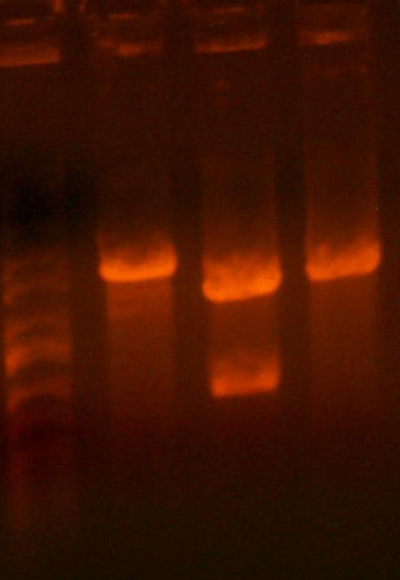
- The lanes are: Marker (Barely came out), 1121, 1122, and rbs pool (cut out)
- 1121 has the right number of bands, but I can't exactly tell if the size of the smaller band is correct.
- 1122 has the wrong number of bands. I'm not that experienced with picking out when a band is undigested plasmid, but even if that were the case for the third band, the smallest band looks much too small.
- Anyway, I cut out the smallest bands from 1121 and 1122 and stored them for tomorrow. Maybe Chris will have something to add to my gelatinous adventures.
198.128.27.101 13:55, 25 May 2007 (PDT)
- Preliminary ranking of RBS's picked from the library based on culture colors: (Bca1101 - Bca1128's in TG1)
Stronger --> Weaker
24, 1, 10, 18, 2, 3, 5, 9, 19, 4, 12, 11, 20, 21, 16, 13, 17, 7, 6, 15, 22, 14, 23
The first 7 are significantly more red than the rest.
# 11 was lost due to a tragic mini prepping accident. May it rest in peace.
AustinDay 16:38, 3 June 2007 (EDT)
- Time to start some awesomeness.
- Biobrick numbers: 051-100 Austin Day
- The next few entries are cut and pasted from my Arkin Wiki because I started making entries on that before this wiki was set up.